Analysis of circulating tumor DNA during checkpoint inhibition in metastatic melanoma using a tumor-agnostic panel
- PMID: 37294123
- PMCID: PMC10470440
- DOI: 10.1097/CMR.0000000000000903
Analysis of circulating tumor DNA during checkpoint inhibition in metastatic melanoma using a tumor-agnostic panel
Abstract
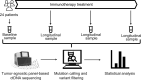
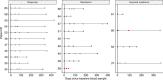
Immunotherapy has revolutionized treatment of patients diagnosed with metastatic melanoma, where nearly half of patients receive clinical benefit. However, immunotherapy is also associated with immune-related adverse events, which may be severe and persistent. It is therefore important to identify patients that do not benefit from therapy early. Currently, regularly scheduled CT scans are used to investigate size changes in target lesions to evaluate progression and therapy response. This study aims to explore if panel-based analysis of circulating tumor DNA (ctDNA) taken at 3-week intervals may provide a window into the growing cancer, can be used to identify nonresponding patients early, and determine genomic alterations associated with acquired resistance to checkpoint immunotherapy without analysis of tumor tissue biopsies. We designed a gene panel for ctDNA analysis and sequenced 4-6 serial plasma samples from 24 patients with unresectable stage III or IV melanoma and treated with first-line checkpoint inhibitors enrolled at the Department of Oncology at Aarhus University Hospital, Denmark. TERT was the most mutated gene found in ctDNA and associated with a poor prognosis. We detected more ctDNA in patients with high metastatic load, which indicates that more aggressive tumors release more ctDNA into the bloodstream. Although we did not find evidence of specific mutations associated with acquired resistance, we did demonstrate in this limited cohort of 24 patients that untargeted, panel-based ctDNA analysis has the potential to be used as a minimally invasive tool in clinical practice to identify patients where the benefits of immunotherapy outweigh the drawbacks.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
There are no conflicts of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

