The paralogues MAGOH and MAGOHB are oncogenic factors in high-grade gliomas and safeguard the splicing of cell division and cell cycle genes
- PMID: 37294214
- PMCID: PMC10259345
- DOI: 10.1080/15476286.2023.2221511
The paralogues MAGOH and MAGOHB are oncogenic factors in high-grade gliomas and safeguard the splicing of cell division and cell cycle genes
Abstract
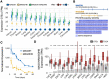
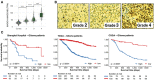
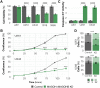
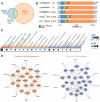
The exon junction complex (EJC) plays key roles throughout the lifespan of RNA and is particularly relevant in the nervous system. We investigated the roles of two EJC members, the paralogs MAGOH and MAGOHB, with respect to brain tumour development. High MAGOH/MAGOHB expression was observed in 14 tumour types; glioblastoma (GBM) showed the greatest difference compared to normal tissue. Increased MAGOH/MAGOHB expression was associated with poor prognosis in glioma patients, while knockdown of MAGOH/MAGOHB affected different cancer phenotypes. Reduced MAGOH/MAGOHB expression in GBM cells caused alterations in the splicing profile, including re-splicing and skipping of multiple exons. The binding profiles of EJC proteins indicated that exons affected by MAGOH/MAGOHB knockdown accumulated fewer complexes on average, providing a possible explanation for their sensitivity to MAGOH/MAGOHB knockdown. Transcripts (genes) showing alterations in the splicing profile are mainly implicated in cell division, cell cycle, splicing, and translation. We propose that high MAGOH/MAGOHB levels are required to safeguard the splicing of genes in high demand in scenarios requiring increased cell proliferation (brain development and GBM growth), ensuring efficient cell division, cell cycle regulation, and gene expression (splicing and translation). Since differentiated neuronal cells do not require increased MAGOH/MAGOHB expression, targeting these paralogs is a potential option for treating GBM.
Keywords: RNA-binding proteins; cancer genomics; gene expression regulation; glioblastoma; glioma; splicing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Multifaceted roles of MAGOH Proteins.Mol Biol Rep. 2023 Feb;50(2):1931-1941. doi: 10.1007/s11033-022-07904-1. Epub 2022 Nov 17. Mol Biol Rep. 2023. PMID: 36396768 Review.
-
MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A.Cells. 2022 Nov 30;11(23):3859. doi: 10.3390/cells11233859. Cells. 2022. PMID: 36497117 Free PMC article.
-
Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay.RNA Biol. 2013 Aug;10(8):1291-8. doi: 10.4161/rna.25827. Epub 2013 Jul 23. RNA Biol. 2013. PMID: 23917022 Free PMC article.
-
Genome-scale analysis identifies paralog lethality as a vulnerability of chromosome 1p loss in cancer.Nat Genet. 2018 Jul;50(7):937-943. doi: 10.1038/s41588-018-0155-3. Epub 2018 Jun 28. Nat Genet. 2018. PMID: 29955178 Free PMC article.
-
The Physiological Roles of the Exon Junction Complex in Development and Diseases.Cells. 2022 Apr 1;11(7):1192. doi: 10.3390/cells11071192. Cells. 2022. PMID: 35406756 Free PMC article. Review.
Cited by
-
MAGOH promotes gastric cancer progression via hnRNPA1 expression inhibition-mediated RONΔ160/PI3K/AKT signaling pathway activation.J Exp Clin Cancer Res. 2024 Jan 25;43(1):32. doi: 10.1186/s13046-024-02946-8. J Exp Clin Cancer Res. 2024. PMID: 38268030 Free PMC article.
-
Integrative bioinformatics and experimental analyses identify U2SURP as a novel lactylation-related prognostic signature in esophageal carcinoma.Immunol Res. 2025 Feb 3;73(1):45. doi: 10.1007/s12026-024-09589-z. Immunol Res. 2025. PMID: 39900790
-
Mago nashi controls auxin-mediated embryo patterning in Arabidopsis by regulating transcript abundance.New Phytol. 2025 Jul;247(1):14-23. doi: 10.1111/nph.70154. Epub 2025 Apr 18. New Phytol. 2025. PMID: 40251862 Free PMC article. No abstract available.
References
-
- Gebauer F, Schwarzl T, Valcárcel J, et al. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3):185–198. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
