Optimized protocol for in vivo affinity purification proteomics and biochemistry using C. elegans
- PMID: 37294631
- PMCID: PMC10323129
- DOI: 10.1016/j.xpro.2023.102262
Optimized protocol for in vivo affinity purification proteomics and biochemistry using C. elegans
Abstract
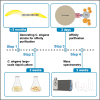
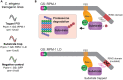
We present an optimized protocol for in vivo affinity purification proteomics and biochemistry using the model organism C. elegans. We describe steps for target tagging, large-scale culture, affinity purification using a cryomill, mass spectrometry and validation of candidate binding proteins. Our approach has proven successful for identifying protein-protein interactions and signaling networks with verified functional relevance. Our protocol is also suitable for biochemical evaluation of protein-protein interactions in vivo. For complete details on the use and execution of this protocol, please refer to Crawley et al.,1 Giles et al.,2 and Desbois et al.3.
Keywords: Mass Spectrometry; Model Organisms; Neuroscience; Protein Biochemistry; Protein expression and purification; Proteomics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures








References
Grants and funding
LinkOut - more resources
Full Text Sources

