Chromosome-level genome assembly of Hydractinia symbiolongicarpus
- PMID: 37294738
- PMCID: PMC10411563
- DOI: 10.1093/g3journal/jkad107
Chromosome-level genome assembly of Hydractinia symbiolongicarpus
Abstract
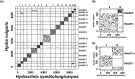
Hydractinia symbiolongicarpus is a pioneering model organism for stem cell biology, being one of only a few animals with adult pluripotent stem cells (known as i-cells). However, the unavailability of a chromosome-level genome assembly has hindered a comprehensive understanding of global gene regulatory mechanisms underlying the function and evolution of i-cells. Here, we report the first chromosome-level genome assembly of H. symbiolongicarpus (HSymV2.0) using PacBio HiFi long-read sequencing and Hi-C scaffolding. The final assembly is 483 Mb in total length with 15 chromosomes representing 99.8% of the assembly. Repetitive sequences were found to account for 296 Mb (61%) of the total genome; we provide evidence for at least two periods of repeat expansion in the past. A total of 25,825 protein-coding genes were predicted in this assembly, which include 93.1% of the metazoan Benchmarking Universal Single-Copy Orthologs (BUSCO) gene set. 92.8% (23,971 genes) of the predicted proteins were functionally annotated. The H. symbiolongicarpus genome showed a high degree of macrosynteny conservation with the Hydra vulgaris genome. This chromosome-level genome assembly of H. symbiolongicarpus will be an invaluable resource for the research community that enhances broad biological studies on this unique model organism.
Keywords: Hydractinia; HSymV2.0; chromosome-level genome assembly; hydrozoa; stem cell.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest statement The authors declare that they have no conflicts of interest.
Figures


References
-
- Bosch TCG, David CN. Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol. 1987;121(1):182–191. doi: 10.1016/0012-1606(87)90151-5. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
