Distinctive microbial community and genome structure in coastal seawater from a human-made port and nearby offshore island in northern Taiwan facing the Northwestern Pacific Ocean
- PMID: 37294811
- PMCID: PMC10256201
- DOI: 10.1371/journal.pone.0284022
Distinctive microbial community and genome structure in coastal seawater from a human-made port and nearby offshore island in northern Taiwan facing the Northwestern Pacific Ocean
Abstract
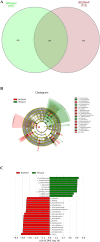
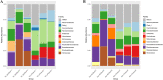
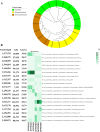
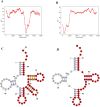
Pollution in human-made fishing ports caused by petroleum from boats, dead fish, toxic chemicals, and effluent poses a challenge to the organisms in seawater. To decipher the impact of pollution on the microbiome, we collected surface water from a fishing port and a nearby offshore island in northern Taiwan facing the Northwestern Pacific Ocean. By employing 16S rRNA gene amplicon sequencing and whole-genome shotgun sequencing, we discovered that Rhodobacteraceae, Vibrionaceae, and Oceanospirillaceae emerged as the dominant species in the fishing port, where we found many genes harboring the functions of antibiotic resistance (ansamycin, nitroimidazole, and aminocoumarin), metal tolerance (copper, chromium, iron and multimetal), virulence factors (chemotaxis, flagella, T3SS1), carbohydrate metabolism (biofilm formation and remodeling of bacterial cell walls), nitrogen metabolism (denitrification, N2 fixation, and ammonium assimilation), and ABC transporters (phosphate, lipopolysaccharide, and branched-chain amino acids). The dominant bacteria at the nearby offshore island (Alteromonadaceae, Cryomorphaceae, Flavobacteriaceae, Litoricolaceae, and Rhodobacteraceae) were partly similar to those in the South China Sea and the East China Sea. Furthermore, we inferred that the microbial community network of the cooccurrence of dominant bacteria on the offshore island was connected to dominant bacteria in the fishing port by mutual exclusion. By examining the assembled microbial genomes collected from the coastal seawater of the fishing port, we revealed four genomic islands containing large gene-containing sequences, including phage integrase, DNA invertase, restriction enzyme, DNA gyrase inhibitor, and antitoxin HigA-1. In this study, we provided clues for the possibility of genomic islands as the units of horizontal transfer and as the tools of microbes for facilitating adaptation in a human-made port environment.
Copyright: © 2023 Shih et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Rhodobacteraceae are Prevalent and Ecologically Crucial Bacterial Members in Marine Biofloc Aquaculture.J Microbiol. 2024 Nov;62(11):985-997. doi: 10.1007/s12275-024-00187-0. Epub 2024 Nov 15. J Microbiol. 2024. PMID: 39546167
-
Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater.FEMS Microbiol Ecol. 2013 Aug;85(2):348-57. doi: 10.1111/1574-6941.12122. Epub 2013 Apr 18. FEMS Microbiol Ecol. 2013. PMID: 23551015
-
Culture dependent and independent analysis and appraisal of early stage biofilm-forming bacterial community composition in the Southern coastal seawater of India.Sci Total Environ. 2019 May 20;666:308-320. doi: 10.1016/j.scitotenv.2019.02.171. Epub 2019 Feb 12. Sci Total Environ. 2019. PMID: 30798240
-
Spatial and vertical distribution of bacterial community in the northern South China Sea.Ecotoxicology. 2015 Oct;24(7-8):1478-85. doi: 10.1007/s10646-015-1472-2. Epub 2015 May 9. Ecotoxicology. 2015. PMID: 25956981
-
Genome analysis of Rubritalea profundi SAORIC-165T, the first deep-sea verrucomicrobial isolate, from the northwestern Pacific Ocean.J Microbiol. 2019 May;57(5):413-422. doi: 10.1007/s12275-019-8712-8. Epub 2019 Feb 26. J Microbiol. 2019. PMID: 30806980
Cited by
-
Iodate reduction by marine aerobic bacteria.Front Microbiol. 2024 Sep 18;15:1446596. doi: 10.3389/fmicb.2024.1446596. eCollection 2024. Front Microbiol. 2024. PMID: 39360326 Free PMC article.
References
-
- Chen J, McIlroy SE, Archana A, Baker DM, Panagiotou G. A pollution gradient contributes to the taxonomic, functional, and resistome diversity of microbial communities in marine sediments. Microbiome. 2019;7(1):104. Epub 2019/07/17. doi: 10.1186/s40168-019-0714-6 ; PubMed Central PMCID: PMC6632204. - DOI - PMC - PubMed
-
- Miettinen H, Bomberg M, Nyyssonen M, Reunamo A, Jorgensen KS, Vikman M. Oil degradation potential of microbial communities in water and sediment of Baltic Sea coastal area. PLoS One. 2019;14(7):e0218834. Epub 2019/07/03. doi: 10.1371/journal.pone.0218834 ; PubMed Central PMCID: PMC6605675. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

