Biofilm formation on human immune cells is a multicellular predation strategy of Vibrio cholerae
- PMID: 37295405
- PMCID: PMC10256282
- DOI: 10.1016/j.cell.2023.05.008
Biofilm formation on human immune cells is a multicellular predation strategy of Vibrio cholerae
Abstract
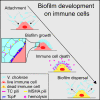
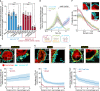
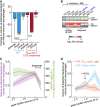
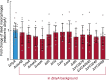
Biofilm formation is generally recognized as a bacterial defense mechanism against environmental threats, including antibiotics, bacteriophages, and leukocytes of the human immune system. Here, we show that for the human pathogen Vibrio cholerae, biofilm formation is not only a protective trait but also an aggressive trait to collectively predate different immune cells. We find that V. cholerae forms biofilms on the eukaryotic cell surface using an extracellular matrix comprising primarily mannose-sensitive hemagglutinin pili, toxin-coregulated pili, and the secreted colonization factor TcpF, which differs from the matrix composition of biofilms on other surfaces. These biofilms encase immune cells and establish a high local concentration of a secreted hemolysin to kill the immune cells before the biofilms disperse in a c-di-GMP-dependent manner. Together, these results uncover how bacteria employ biofilm formation as a multicellular strategy to invert the typical relationship between human immune cells as the hunters and bacteria as the hunted.
Keywords: cholera infection; enteroid; host-pathogen interaction; immunity; organoid; type IV pili.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests All authors declare no competing interests.
Figures






Comment in
-
Bacteria form unique biofilms to kill immune cells.Nat Rev Immunol. 2023 Aug;23(8):474. doi: 10.1038/s41577-023-00914-5. Nat Rev Immunol. 2023. PMID: 37402991 No abstract available.
-
The Vibrio cholerae maneuver.Trends Immunol. 2023 Aug;44(8):565-567. doi: 10.1016/j.it.2023.06.009. Epub 2023 Jul 8. Trends Immunol. 2023. PMID: 37429798 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

