Whole-Exome Sequencing Identifies Homozygote Nonsense Variants in LMOD2 Gene Causing Infantile Dilated Cardiomyopathy
- PMID: 37296576
- PMCID: PMC10252268
- DOI: 10.3390/cells12111455
Whole-Exome Sequencing Identifies Homozygote Nonsense Variants in LMOD2 Gene Causing Infantile Dilated Cardiomyopathy
Abstract
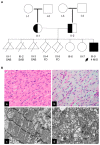
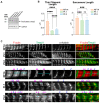
As an essential component of the sarcomere, actin thin filament stems from the Z-disk extend toward the middle of the sarcomere and overlaps with myosin thick filaments. Elongation of the cardiac thin filament is essential for normal sarcomere maturation and heart function. This process is regulated by the actin-binding proteins Leiomodins (LMODs), among which LMOD2 has recently been identified as a key regulator of thin filament elongation to reach a mature length. Few reports have implicated homozygous loss of function variants of LMOD2 in neonatal dilated cardiomyopathy (DCM) associated with thin filament shortening. We present the fifth case of DCM due to biallelic variants in the LMOD2 gene and the second case with the c.1193G>A (p.W398*) nonsense variant identified by whole-exome sequencing. The proband is a 4-month male infant of Hispanic descent with advanced heart failure. Consistent with previous reports, a myocardial biopsy exhibited remarkably short thin filaments. However, compared to other cases of identical or similar biallelic variants, the patient presented here has an unusually late onset of cardiomyopathy during infancy. Herein, we present the phenotypic and histological features of this variant, confirm the pathogenic impact on protein expression and sarcomere structure, and discuss the current knowledge of LMOD2-related cardiomyopathy.
Keywords: DCM; LMOD2; heart maturation; leiomodins; neonatal cardiomyopathy; sarcomere; thin filament; whole-exome sequencing; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Pappas C.T., Mayfield R.M., Henderson C., Jamilpour N., Cover C., Hernandez Z., Hutchinson K.R., Chu M., Nam K.-H., Valdez J.M., et al. Knockout of Lmod2 Results in Shorter Thin Filaments Followed by Dilated Cardiomyopathy and Juvenile Lethality. Proc. Natl. Acad. Sci. USA. 2015;112:13573–13578. doi: 10.1073/pnas.1508273112. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

