scViewer: An Interactive Single-Cell Gene Expression Visualization Tool
- PMID: 37296611
- PMCID: PMC10253102
- DOI: 10.3390/cells12111489
scViewer: An Interactive Single-Cell Gene Expression Visualization Tool
Abstract
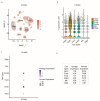
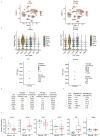
Single-cell RNA sequencing (scRNA-seq) is an attractive technology for researchers to gain valuable insights into the cellular processes and cell type diversity present in all tissues. The data generated by the scRNA-seq experiment are high-dimensional and complex in nature. Several tools are now available to analyze the raw scRNA-seq data from public databases; however, simple and easy-to-explore single-cell gene expression visualization tools focusing on differential expression and co-expression are lacking. Here, we present scViewer, an interactive graphical user interface (GUI) R/Shiny application designed to facilitate the visualization of scRNA-seq gene expression data. With the processed Seurat RDS object as input, scViewer utilizes several statistical approaches to provide detailed information on the loaded scRNA-seq experiment and generates publication-ready plots. The major functionalities of scViewer include exploring cell-type-specific gene expression, co-expression analysis of two genes, and differential expression analysis with different biological conditions considering both cell-level and subject-level variations using negative binomial mixed modeling. We utilized a publicly available dataset (brain cells from a study of Alzheimer's disease to demonstrate the utility of our tool. scViewer can be downloaded from GitHub as a Shiny app with local installation. Overall, scViewer is a user-friendly application that will allow researchers to visualize and interpret the scRNA-seq data efficiently for multi-condition comparison by performing gene-level differential expression and co-expression analysis on the fly. Considering the functionalities of this Shiny app, scViewer can be a great resource for collaboration between bioinformaticians and wet lab scientists for faster data visualizations.
Keywords: R Shiny; bioinformatics; co-expression; differential expression analysis; gene expression; scRNA-seq; single-cell RNA sequencing.
Conflict of interest statement
Teva Pharmaceuticals Industries Ltd. develops, produces, and markets affordable, high-quality generic drugs and specialty pharmaceuticals. At the time of the study, all the authors were employed by Teva Pharmaceutical Industries Ltd. The authors declare the absence of any potential conflicts of interest, such as commercial or financial relationships, pertaining to the conduction of this study.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

