In Situ Identification of Both IL-4 and IL-10 Cytokine-Receptor Interactions during Tissue Regeneration
- PMID: 37296643
- PMCID: PMC10253026
- DOI: 10.3390/cells12111522
In Situ Identification of Both IL-4 and IL-10 Cytokine-Receptor Interactions during Tissue Regeneration
Abstract
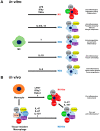
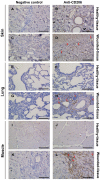
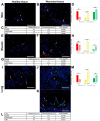
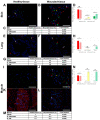
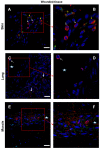
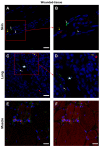
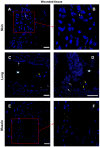
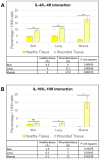
Cytokines secreted by individual immune cells regulate tissue regeneration and allow communication between various cell types. Cytokines bind to cognate receptors and trigger the healing process. Determining the orchestration of cytokine interactions with their receptors on their cellular targets is essential to fully understanding the process of inflammation and tissue regeneration. To this end, we have investigated the interactions of Interleukin-4 cytokine (IL-4)/Interleukin-4 cytokine receptor (IL-4R) and Interleukin-10 cytokine (IL-10)/Interleukin-10 cytokine receptor (IL-10R) using in situ Proximity Ligation Assays in a regenerative model of skin, muscle and lung tissues in the mini-pig. The pattern of protein-protein interactions was distinct for the two cytokines. IL-4 bound predominantly to receptors on macrophages and endothelial cells around the blood vessels while the target cells of IL-10 were mainly receptors on muscle cells. Our results show that in situ studies of cytokine-receptor interactions can unravel the fine details of the mechanism of action of cytokines.
Keywords: IL-10; IL-4; interaction; proximity ligation assay; receptor; regeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

