A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer
- PMID: 37296997
- PMCID: PMC10252086
- DOI: 10.3390/cancers15113035
A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer
Abstract
Background: Molecular biomarkers that predict disease progression can help identify tumor subtypes and shape treatment plans. In this study, we aimed to identify robust biomarkers of prognosis in gastric cancer based on transcriptomic data obtained from primary gastric tumors.
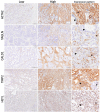
Methods: Microarray, RNA sequencing, and single-cell RNA sequencing-based gene expression data from gastric tumors were obtained from public databases. Freshly frozen gastric tumors (n = 42) and matched FFPE (formalin-fixed, paraffin-embedded) (n = 40) tissues from a Turkish gastric cancer cohort were used for quantitative real-time PCR and immunohistochemistry-based assessments of gene expression, respectively.
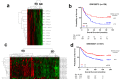
Results: A novel list of 20 prognostic genes was identified and used for the classification of gastric tumors into two major tumor subgroups with differential stromal gene expression ("Stromal-UP" (SU) and "Stromal-DOWN" (SD)). The SU group had a more mesenchymal profile with an enrichment of extracellular matrix-related gene sets and a poor prognosis compared to the SD group. Expression of the genes within the signature correlated with the expression of mesenchymal markers ex vivo. A higher stromal content in FFPE tissues was associated with shorter overall survival.
Conclusions: A stroma-rich, mesenchymal subgroup among gastric tumors identifies an unfavorable clinical outcome in all cohorts tested.
Keywords: biomarker; gastric cancer; prognosis; stroma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Marano L., D’Ignazio A., Cammillini F., Angotti R., Messina M., Marrelli D., Roviello F. Comparison between 7th and 8th edition of AJCC TNM staging system for gastric cancer: Old problems and new perspectives. Transl. Gastroenterol. Hepatol. 2019;4:22. doi: 10.21037/tgh.2019.03.09. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

