Glioblastoma Metabolism: Insights and Therapeutic Strategies
- PMID: 37298093
- PMCID: PMC10252397
- DOI: 10.3390/ijms24119137
Glioblastoma Metabolism: Insights and Therapeutic Strategies
Abstract
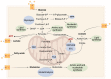
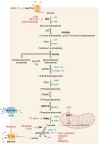
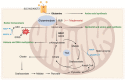
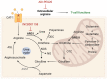
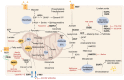
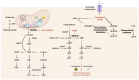
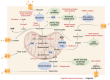
Tumor metabolism is emerging as a potential target for cancer therapies. This new approach holds particular promise for the treatment of glioblastoma, a highly lethal brain tumor that is resistant to conventional treatments, for which improving therapeutic strategies is a major challenge. The presence of glioma stem cells is a critical factor in therapy resistance, thus making it essential to eliminate these cells for the long-term survival of cancer patients. Recent advancements in our understanding of cancer metabolism have shown that glioblastoma metabolism is highly heterogeneous, and that cancer stem cells exhibit specific metabolic traits that support their unique functionality. The objective of this review is to examine the metabolic changes in glioblastoma and investigate the role of specific metabolic processes in tumorigenesis, as well as associated therapeutic approaches, with a particular focus on glioma stem cell populations.
Keywords: cancer stem cells; glioblastoma; metabolism; therapy; tumorigenic processes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells.Biochim Biophys Acta Rev Cancer. 2018 Apr;1869(2):175-188. doi: 10.1016/j.bbcan.2018.01.004. Epub 2018 Jan 31. Biochim Biophys Acta Rev Cancer. 2018. PMID: 29378228 Free PMC article. Review.
-
Dedifferentiation of Glioma Cells to Glioma Stem-like Cells By Therapeutic Stress-induced HIF Signaling in the Recurrent GBM Model.Mol Cancer Ther. 2016 Dec;15(12):3064-3076. doi: 10.1158/1535-7163.MCT-15-0675. Epub 2016 Oct 7. Mol Cancer Ther. 2016. PMID: 27765847 Free PMC article.
-
Targeting Folate Metabolism Is Selectively Cytotoxic to Glioma Stem Cells and Effectively Cooperates with Differentiation Therapy to Eliminate Tumor-Initiating Cells in Glioma Xenografts.Int J Mol Sci. 2021 Oct 27;22(21):11633. doi: 10.3390/ijms222111633. Int J Mol Sci. 2021. PMID: 34769063 Free PMC article.
-
Cancer stem cells in glioblastoma--molecular signaling and therapeutic targeting.Protein Cell. 2010 Jul;1(7):638-55. doi: 10.1007/s13238-010-0078-y. Epub 2010 Jul 29. Protein Cell. 2010. PMID: 21203936 Free PMC article. Review.
-
Cancer stem cells: The potential role of autophagy, proteolysis, and cathepsins in glioblastoma stem cells.Tumour Biol. 2017 Mar;39(3):1010428317692227. doi: 10.1177/1010428317692227. Tumour Biol. 2017. PMID: 28347245 Review.
Cited by
-
Mitochondrial Protein Density, Biomass, and Bioenergetics as Predictors for the Efficacy of Glioma Treatments.Int J Mol Sci. 2024 Jun 27;25(13):7038. doi: 10.3390/ijms25137038. Int J Mol Sci. 2024. PMID: 39000148 Free PMC article.
-
A molecular signature for the G6PC3/SLC37A2/SLC37A4 interactors in glioblastoma disease progression and in the acquisition of a brain cancer stem cell phenotype.Front Endocrinol (Lausanne). 2023 Nov 16;14:1265698. doi: 10.3389/fendo.2023.1265698. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 38034009 Free PMC article.
-
Metabolic shifts in glioblastoma: unraveling altered pathways and exploring novel therapeutic avenues.Mol Biol Rep. 2025 Jan 22;52(1):146. doi: 10.1007/s11033-025-10242-7. Mol Biol Rep. 2025. PMID: 39841290 Review.
-
Metabolic Roles of HIF1, c-Myc, and p53 in Glioma Cells.Metabolites. 2024 Apr 25;14(5):249. doi: 10.3390/metabo14050249. Metabolites. 2024. PMID: 38786726 Free PMC article. Review.
-
Investigative needle core biopsies support multimodal deep-data generation in glioblastoma.Nat Commun. 2025 Apr 28;16(1):3957. doi: 10.1038/s41467-025-58452-8. Nat Commun. 2025. PMID: 40295505 Free PMC article.
References
-
- Verhaak R.G.W., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. - DOI - PMC - PubMed
-
- Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., DeCarvalho A.C., Lyu S., Li P., Li Y., et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

