Emerging Role of DREAM in Healthy Brain and Neurological Diseases
- PMID: 37298129
- PMCID: PMC10252307
- DOI: 10.3390/ijms24119177
Emerging Role of DREAM in Healthy Brain and Neurological Diseases
Abstract
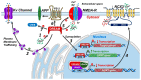
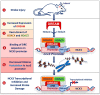
The downstream regulatory element antagonist modulator (DREAM) is a multifunctional Ca2+-sensitive protein exerting a dual mechanism of action to regulate several Ca2+-dependent processes. Upon sumoylation, DREAM enters in nucleus where it downregulates the expression of several genes provided with a consensus sequence named dream regulatory element (DRE). On the other hand, DREAM could also directly modulate the activity or the localization of several cytosolic and plasma membrane proteins. In this review, we summarize recent advances in the knowledge of DREAM dysregulation and DREAM-dependent epigenetic remodeling as a central mechanism in the progression of several diseases affecting central nervous system, including stroke, Alzheimer's and Huntington's diseases, amyotrophic lateral sclerosis, and neuropathic pain. Interestingly, DREAM seems to exert a common detrimental role in these diseases by inhibiting the transcription of several neuroprotective genes, including the sodium/calcium exchanger isoform 3 (NCX3), brain-derived neurotrophic factor (BDNF), pro-dynorphin, and c-fos. These findings lead to the concept that DREAM might represent a pharmacological target to ameliorate symptoms and reduce neurodegenerative processes in several pathological conditions affecting central nervous system.
Keywords: DREAM; KCNIP3; calsenilin; neurodegeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

