Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion
- PMID: 37298367
- PMCID: PMC10253471
- DOI: 10.3390/ijms24119415
Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion
Abstract
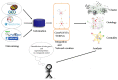
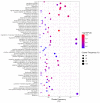
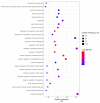
Complex diseases are associated with the effects of multiple genes, proteins, and biological pathways. In this context, the tools of Network Medicine are compatible as a platform to systematically explore not only the molecular complexity of a specific disease but may also lead to the identification of disease modules and pathways. Such an approach enables us to gain a better understanding of how environmental chemical exposures affect the function of human cells, providing better perceptions about the mechanisms involved and helping to monitor/prevent exposure and disease to chemicals such as benzene and malathion. We selected differentially expressed genes for exposure to benzene and malathion. The construction of interaction networks was carried out using GeneMANIA and STRING. Topological properties were calculated using MCODE, BiNGO, and CentiScaPe, and a Benzene network composed of 114 genes and 2415 interactions was obtained. After topological analysis, five networks were identified. In these subnets, the most interconnected nodes were identified as: IL-8, KLF6, KLF4, JUN, SERTAD1, and MT1H. In the Malathion network, composed of 67 proteins and 134 interactions, HRAS and STAT3 were the most interconnected nodes. Path analysis, combined with various types of high-throughput data, reflects biological processes more clearly and comprehensively than analyses involving the evaluation of individual genes. We emphasize the central roles played by several important hub genes obtained by exposure to benzene and malathion.
Keywords: benzene; malathion; network medicine; occupational health.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Chen L., Wang R.-S., Zhang X. Biomolecular Networks: Methods and Applications in Systems Biology. 1st ed. Wiley; Hoboken, NJ, USA: 2009. p. 387.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

