Secondary Structures of MERS-CoV, SARS-CoV, and SARS-CoV-2 Spike Proteins Revealed by Infrared Vibrational Spectroscopy
- PMID: 37298500
- PMCID: PMC10253540
- DOI: 10.3390/ijms24119550
Secondary Structures of MERS-CoV, SARS-CoV, and SARS-CoV-2 Spike Proteins Revealed by Infrared Vibrational Spectroscopy
Abstract
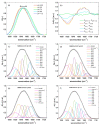
All coronaviruses are characterized by spike glycoproteins whose S1 subunits contain the receptor binding domain (RBD). The RBD anchors the virus to the host cellular membrane to regulate the virus transmissibility and infectious process. Although the protein/receptor interaction mainly depends on the spike's conformation, particularly on its S1 unit, their secondary structures are poorly known. In this paper, the S1 conformation was investigated for MERS-CoV, SARS-CoV, and SARS-CoV-2 at serological pH by measuring their Amide I infrared absorption bands. The SARS-CoV-2 S1 secondary structure revealed a strong difference compared to those of MERS-CoV and SARS-CoV, with a significant presence of extended β-sheets. Furthermore, the conformation of the SARS-CoV-2 S1 showed a significant change by moving from serological pH to mild acidic and alkaline pH conditions. Both results suggest the capability of infrared spectroscopy to follow the secondary structure adaptation of the SARS-CoV-2 S1 to different environments.
Keywords: ATR-IR spectroscopy; MERS-CoV; SARS-CoV; SARS-CoV-2; conformation; secondary structure; spike glycoproteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

