GmUFO1 Regulates Floral Organ Number and Shape in Soybean
- PMID: 37298613
- PMCID: PMC10253600
- DOI: 10.3390/ijms24119662
GmUFO1 Regulates Floral Organ Number and Shape in Soybean
Abstract
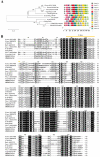
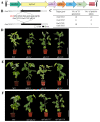
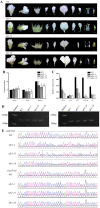
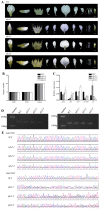
The UNUSUAL FLORAL ORGANS (UFO) gene is an essential regulatory factor of class B genes and plays a vital role in the process of inflorescence primordial and flower primordial development. The role of UFO genes in soybean was investigated to better understand the development of floral organs through gene cloning, expression analysis, and gene knockout. There are two copies of UFO genes in soybean and in situ hybridization, which have demonstrated similar expression patterns of the GmUFO1 and GmUFO2 genes in the flower primordium. The phenotypic observation of GmUFO1 knockout mutant lines (Gmufo1) showed an obvious alteration in the floral organ number and shape and mosaic organ formation. By contrast, GmUFO2 knockout mutant lines (Gmufo2) showed no obvious difference in the floral organs. However, the GmUFO1 and GmUFO2 double knockout lines (Gmufo1ufo2) showed more mosaic organs than the Gmufo1 lines, in addition to the alteration in the organ number and shape. Gene expression analysis also showed differences in the expression of major ABC function genes in the knockout lines. Based on the phenotypic and expression analysis, our results suggest the major role of GmUFO1 in the regulation of flower organ formation in soybeans and that GmUFO2 does not have any direct effect but might have an interaction role with GmUFO1 in the regulation of flower development. In conclusion, the present study identified UFO genes in soybean and improved our understanding of floral development, which could be useful for flower designs in hybrid soybean breeding.
Keywords: GmUFOs; floral organ number; floral organ shape; knockout; soybean.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression.Planta. 2006 Mar;223(4):769-78. doi: 10.1007/s00425-005-0138-3. Epub 2005 Oct 22. Planta. 2006. PMID: 16244866
-
UFO: an Arabidopsis gene involved in both floral meristem and floral organ development.Plant Cell. 1995 May;7(5):529-48. doi: 10.1105/tpc.7.5.529. Plant Cell. 1995. PMID: 7780306 Free PMC article.
-
A soybean MADS-box protein modulates floral organ numbers, petal identity and sterility.BMC Plant Biol. 2014 Apr 2;14:89. doi: 10.1186/1471-2229-14-89. BMC Plant Biol. 2014. PMID: 24693922 Free PMC article.
-
Molecular Mechanisms of Floral Boundary Formation in Arabidopsis.Int J Mol Sci. 2016 Mar 2;17(3):317. doi: 10.3390/ijms17030317. Int J Mol Sci. 2016. PMID: 26950117 Free PMC article. Review.
-
Specification of floral organs in Arabidopsis.J Exp Bot. 2014 Jan;65(1):1-9. doi: 10.1093/jxb/ert385. Epub 2013 Nov 25. J Exp Bot. 2014. PMID: 24277279 Review.
References
-
- Thomson B., Wellmer F. Molecular regulation of flower development. Curr. Top. Dev. Biol. 2019;131:185–210. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

