The Combination of a Human Biomimetic Liver Microphysiology System with BIOLOGXsym, a Quantitative Systems Toxicology (QST) Modeling Platform for Macromolecules, Provides Mechanistic Understanding of Tocilizumab- and GGF2-Induced Liver Injury
- PMID: 37298645
- PMCID: PMC10253699
- DOI: 10.3390/ijms24119692
The Combination of a Human Biomimetic Liver Microphysiology System with BIOLOGXsym, a Quantitative Systems Toxicology (QST) Modeling Platform for Macromolecules, Provides Mechanistic Understanding of Tocilizumab- and GGF2-Induced Liver Injury
Abstract
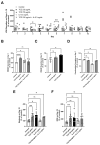
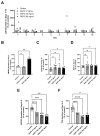
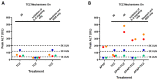
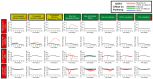
Biologics address a range of unmet clinical needs, but the occurrence of biologics-induced liver injury remains a major challenge. Development of cimaglermin alfa (GGF2) was terminated due to transient elevations in serum aminotransferases and total bilirubin. Tocilizumab has been reported to induce transient aminotransferase elevations, requiring frequent monitoring. To evaluate the clinical risk of biologics-induced liver injury, a novel quantitative systems toxicology modeling platform, BIOLOGXsym™, representing relevant liver biochemistry and the mechanistic effects of biologics on liver pathophysiology, was developed in conjunction with clinically relevant data from a human biomimetic liver microphysiology system. Phenotypic and mechanistic toxicity data and metabolomics analysis from the Liver Acinus Microphysiology System showed that tocilizumab and GGF2 increased high mobility group box 1, indicating hepatic injury and stress. Tocilizumab exposure was associated with increased oxidative stress and extracellular/tissue remodeling, and GGF2 decreased bile acid secretion. BIOLOGXsym simulations, leveraging the in vivo exposure predicted by physiologically-based pharmacokinetic modeling and mechanistic toxicity data from the Liver Acinus Microphysiology System, reproduced the clinically observed liver signals of tocilizumab and GGF2, demonstrating that mechanistic toxicity data from microphysiology systems can be successfully integrated into a quantitative systems toxicology model to identify liabilities of biologics-induced liver injury and provide mechanistic insights into observed liver safety signals.
Keywords: hepatotoxicity; human liver microphysiology system; macromolecule; quantitative systems toxicology (QST) modeling.
Conflict of interest statement
James J. Beaudoin, Lara Clemens, Christina Battista, Lisl K. M. Shoda, Scott Q. Siler, Brett A. Howell, and Kyunghee Yang are employees of Simulations Plus Inc., which received a National Institutes of Health Small Business Innovation Research Award in collaboration with the University of Pittsburgh Drug Discovery Institute to develop a quantitative systems toxicology platform to predict biologics-induced liver injury. Fatima Zaidi, Priya Ramamoorthy, Kari Wong, Rangaprasad Sarangarajan are employees of Metabolon Inc., which provided in-kind support for metabolomics analysis.
Figures





References
-
- FDA . What Are “Biologics” Questions and Answers. US FDA; Silver Spring, MD, USA: 2018.
-
- Opportunities and Challenges in Biologic Drug Delivery. 2017. [(accessed on 4 March 2020)]. Available online: http://www.americanpharmaceuticalreview.com/Featured-Articles/345540-Opp...
-
- Mosedale M., Button D., Jackson J.P., Freeman K.M., Brouwer K.R., Caggiano A.O., Eisen A., Iaci J.F., Parry T.J., Stanulis R., et al. Transient Changes in Hepatic Physiology That Alter Bilirubin and Bile Acid Transport May Explain Elevations in Liver Chemistries Observed in Clinical Trials of GGF2 (Cimaglermin Alfa) Toxicol. Sci. 2018;161:401–411. doi: 10.1093/toxsci/kfx222. - DOI - PubMed
-
- Longo D.M., Generaux G.T., Howell B.A., Siler S.Q., Antoine D.J., Button D., Caggiano A., Eisen A., Iaci J., Stanulis R., et al. Refining Liver Safety Risk Assessment: Application of Mechanistic Modeling and Serum Biomarkers to Cimaglermin Alfa (GGF2) Clinical Trials. Clin. Pharmacol. Ther. 2017;102:961–969. doi: 10.1002/cpt.711. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

