Group I Intron as a Potential Target for Antifungal Compounds: Development of a Trans-Splicing High-Throughput Screening Strategy
- PMID: 37298936
- PMCID: PMC10254303
- DOI: 10.3390/molecules28114460
Group I Intron as a Potential Target for Antifungal Compounds: Development of a Trans-Splicing High-Throughput Screening Strategy
Abstract
The search for safe and efficient new antifungal compounds for agriculture has led to more efforts in finding new modes of action. This involves the discovery of new molecular targets, including coding and non-coding RNA. Rarely found in plants and animals but present in fungi, group I introns are of interest as their complex tertiary structure may allow selective targeting using small molecules. In this work, we demonstrate that group I introns present in phytopathogenic fungi have a self-splicing activity in vitro that can be adapted in a high-throughput screening to find new antifungal compounds. Ten candidate introns from different filamentous fungi were tested and one group ID intron found in F. oxysporum showed high self-splicing efficiency in vitro. We designed the Fusarium intron to act as a trans-acting ribozyme and used a fluorescence-based reporter system to monitor its real time splicing activity. Together, these results are opening the way to study the druggability of such introns in crop pathogen and potentially discover small molecules selectively targeting group I introns in future high-throughput screenings.
Keywords: Fusarium; HTS; RNA; group I; ribozyme; self-splicing.
Conflict of interest statement
The authors are employees of Bayer Crop Science, a leading developer of agricultural seeds and chemistries.
Figures

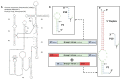


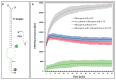
References
-
- Seth P.P., Miyaji A., Jefferson E.A., Sannes-Lowery K.A., Osgood S.A., Propp S.S., Ranken R., Massire C., Sampath R., Ecker D.J., et al. SAR by MS: Discovery of a New Class of RNA-Binding Small Molecules for the Hepatitis C Virus: Internal Ribosome Entry Site IIA Subdomain. J. Med. Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

