Large Artificial microRNA Cluster Genes Confer Effective Resistance against Multiple Tomato Yellow Leaf Curl Viruses in Transgenic Tomato
- PMID: 37299158
- PMCID: PMC10255879
- DOI: 10.3390/plants12112179
Large Artificial microRNA Cluster Genes Confer Effective Resistance against Multiple Tomato Yellow Leaf Curl Viruses in Transgenic Tomato
Abstract
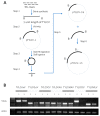
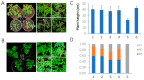
Tomato yellow leaf curl disease (TYLCD) has become the key limiting factor for the production of tomato in many areas because of the continuous infection and recombination of several tomato yellow leaf curl virus (TYLCV)-like species (TYLCLV) which produce novel and destructive viruses. Artificial microRNA (AMIR) is a recent and effective technology used to create viral resistance in major crops. This study applies AMIR technology in two ways, i.e., amiRNA in introns (AMINs) and amiRNA in exons (AMIEs), to express 14 amiRNAs targeting conserved regions in seven TYLCLV genes and their satellite DNA. The resulting pAMIN14 and pAMIE14 vectors can encode large AMIR clusters and their function in silencing reporter genes was validated with transient assays and stable transgenic N. tabacum plants. To assess the efficacy of conferring resistance against TYLCLV, pAMIE14 and pAMIN14 were transformed into tomato cultivar A57 and the resulting transgenic tomato plants were evaluated for their level of resistance to mixed TYLCLV infection. The results suggest that pAMIN14 transgenic lines have a more effective resistance than pAMIE14 transgenic lines, reaching a resistance level comparable to plants carrying the TY1 resistance gene.
Keywords: TY1; artificial miRNA; mixed infection; resistance; tomato yellow leaf curl viruses; whitefly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Silencing AC1 of Tomato leaf curl virus using artificial microRNA confers resistance to leaf curl disease in transgenic tomato.Plant Cell Rep. 2020 Nov;39(11):1565-1579. doi: 10.1007/s00299-020-02584-2. Epub 2020 Aug 29. Plant Cell Rep. 2020. PMID: 32860518
-
Post-transcriptional gene silencing in controlling viruses of the Tomato yellow leaf curl virus complex.Arch Virol. 2006 Dec;151(12):2349-63. doi: 10.1007/s00705-006-0819-7. Epub 2006 Jul 27. Arch Virol. 2006. PMID: 16862387
-
Detection of Satellite DNA Beta in Tomato Plants with Tomato Yellow Leaf Curl Disease in Jordan.Plant Dis. 2014 Jul;98(7):1017. doi: 10.1094/PDIS-02-14-0144-PDN. Plant Dis. 2014. PMID: 30708869
-
Artificial microRNA-mediated resistance against Oman strain of tomato yellow leaf curl virus.Front Plant Sci. 2023 Mar 30;14:1164921. doi: 10.3389/fpls.2023.1164921. eCollection 2023. Front Plant Sci. 2023. PMID: 37063229 Free PMC article.
-
Natural resistance of tomato plants to Tomato yellow leaf curl virus.Front Plant Sci. 2022 Dec 19;13:1081549. doi: 10.3389/fpls.2022.1081549. eCollection 2022. Front Plant Sci. 2022. PMID: 36600922 Free PMC article. Review.
Cited by
-
An Integrative Computational Approach for Identifying Cotton Host Plant MicroRNAs with Potential to Abate CLCuKoV-Bur Infection.Viruses. 2025 Mar 12;17(3):399. doi: 10.3390/v17030399. Viruses. 2025. PMID: 40143327 Free PMC article.
-
Artificial miRNAs and target-mimics as potential tools for crop improvement.Physiol Mol Biol Plants. 2025 Jan;31(1):67-91. doi: 10.1007/s12298-025-01550-0. Epub 2025 Jan 17. Physiol Mol Biol Plants. 2025. PMID: 39901962 Review.
-
microRNAs: Key Regulators in Plant Responses to Abiotic and Biotic Stresses via Endogenous and Cross-Kingdom Mechanisms.Int J Mol Sci. 2024 Jan 18;25(2):1154. doi: 10.3390/ijms25021154. Int J Mol Sci. 2024. PMID: 38256227 Free PMC article. Review.
-
Promising Biotechnological Applications of the Artificial Derivatives Designed and Constructed from Plant microRNA Genes.Plants (Basel). 2025 Jan 22;14(3):325. doi: 10.3390/plants14030325. Plants (Basel). 2025. PMID: 39942887 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

