Expression of RsPORB Is Associated with Radish Root Color
- PMID: 37299194
- PMCID: PMC10255411
- DOI: 10.3390/plants12112214
Expression of RsPORB Is Associated with Radish Root Color
Abstract
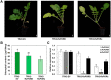
Radish (Raphanus sativus) plants exhibit varied root colors due to the accumulation of chlorophylls and anthocyanins compounds that are beneficial for both human health and visual quality. The mechanisms of chlorophyll biosynthesis have been extensively studied in foliar tissues but remain largely unknown in other tissues. In this study, we examined the role of NADPH:protochlorophyllide oxidoreductases (PORs), which are key enzymes in chlorophyll biosynthesis, in radish roots. The transcript level of RsPORB was abundantly expressed in green roots and positively correlated with chlorophyll content in radish roots. Sequences of the RsPORB coding region were identical between white (948) and green (847) radish breeding lines. Additionally, virus-induced gene silencing assay with RsPORB exhibited reduced chlorophyll contents, verifying that RsPORB is a functional enzyme for chlorophyll biosynthesis. Sequence comparison of RsPORB promoters from white and green radishes showed several insertions and deletions (InDels) and single-nucleotide polymorphisms. Promoter activation assays using radish root protoplasts verified that InDels of the RsPORB promoter contribute to its expression level. These results suggested that RsPORB is one of the key genes underlying chlorophyll biosynthesis and green coloration in non-foliar tissues, such as roots.
Keywords: RsPORB; chlorophyll biosynthesis; green radish; polymorphisms; promoter variation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
RsGLK2.1-RsNF-YA9a module positively regulates the chlorophyll biosynthesis by activating RsHEMA2 in green taproot of radish.Plant Sci. 2023 Sep;334:111768. doi: 10.1016/j.plantsci.2023.111768. Epub 2023 Jun 19. Plant Sci. 2023. PMID: 37343602
-
Molecular mechanism controlling anthocyanin composition and content in radish plants with different root colors.Plant Physiol Biochem. 2023 Nov;204:108091. doi: 10.1016/j.plaphy.2023.108091. Epub 2023 Oct 12. Plant Physiol Biochem. 2023. PMID: 37864927
-
Transcriptome analyses reveal key genes involved in skin color changes of 'Xinlimei' radish taproot.Plant Physiol Biochem. 2019 Jun;139:528-539. doi: 10.1016/j.plaphy.2019.04.006. Epub 2019 Apr 11. Plant Physiol Biochem. 2019. PMID: 31029026
-
Development of Molecular Markers for Predicting Radish (Raphanus sativus) Flesh Color Based on Polymorphisms in the RsTT8 Gene.Plants (Basel). 2021 Jul 6;10(7):1386. doi: 10.3390/plants10071386. Plants (Basel). 2021. PMID: 34371589 Free PMC article.
-
A Comparative Metabolomics Study of Flavonoids in Radish with Different Skin and Flesh Colors (Raphanus sativus L.).J Agric Food Chem. 2020 Dec 9;68(49):14463-14470. doi: 10.1021/acs.jafc.0c05031. Epub 2020 Nov 20. J Agric Food Chem. 2020. PMID: 33216541
Cited by
-
Endonuclease Genes in Rice Are Involved in Phosphate Source Recycling by DNA Decay From Phosphate Deprivation.Physiol Plant. 2025 Jul-Aug;177(4):e70452. doi: 10.1111/ppl.70452. Physiol Plant. 2025. PMID: 40791137 Free PMC article.
References
-
- Fu H., Zeng T., Zhao Y., Luo T., Deng H., Meng C., Luo J., Wang C. Identification of chlorophyll metabolism-and photosynthesis-related genes regulating green flower color in chrysanthemum by integrative transcriptome and weighted correlation network analyses. Genes. 2021;12:449. doi: 10.3390/genes12030449. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

