The Oncogenic Protein Kinase/ATPase RIOK1 Is Up-Regulated via the c-myc/E2F Transcription Factor Axis in Prostate Cancer
- PMID: 37301535
- PMCID: PMC12178385
- DOI: 10.1016/j.ajpath.2023.05.013
The Oncogenic Protein Kinase/ATPase RIOK1 Is Up-Regulated via the c-myc/E2F Transcription Factor Axis in Prostate Cancer
Abstract
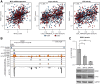
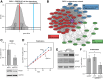
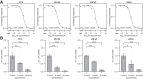
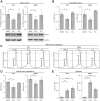
The atypical protein kinase/ATPase RIO kinase (RIOK)-1 is involved in pre-40S ribosomal subunit production, cell-cycle progression, and protein arginine N-methyltransferase 5 methylosome substrate recruitment. RIOK1 overexpression is a characteristic of several malignancies and is correlated with cancer stage, therapy resistance, poor patient survival, and other prognostic factors. However, its role in prostate cancer (PCa) is unknown. In this study, the expression, regulation, and therapeutic potential of RIOK1 in PCa were examined. RIOK1 mRNA and protein expression were elevated in PCa tissue samples and correlated with proliferative and protein homeostasis-related pathways. RIOK1 was identified as a downstream target gene of the c-myc/E2F transcription factors. Proliferation of PCa cells was significantly reduced with RIOK1 knockdown and overexpression of the dominant-negative RIOK1-D324A mutant. Biochemical inhibition of RIOK1 with toyocamycin led to strong antiproliferative effects in androgen receptor-negative and -positive PCa cell lines with EC50 values of 3.5 to 8.8 nmol/L. Rapid decreases in RIOK1 protein expression and total rRNA content, and a shift in the 28S/18S rRNA ratio, were found with toyocamycin treatment. Apoptosis was induced with toyocamycin treatment at a level similar to that with the chemotherapeutic drug docetaxel used in clinical practice. In summary, the current study indicates that RIOK1 is a part of the MYC oncogene network, and as such, could be considered for future treatment of patients with PCa.
Copyright © 2023 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Increased GPR35 expression is correlated with poor prognosis in prostate cancer.World J Surg Oncol. 2025 Jun 18;23(1):239. doi: 10.1186/s12957-025-03893-0. World J Surg Oncol. 2025. PMID: 40533764 Free PMC article.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Cost-effectiveness of enzalutamide with androgen-deprivation therapy (ADT) versus ADT alone for the treatment of high-risk biochemically recurrent non-metastatic castration-sensitive prostate cancer in Canada.J Med Econ. 2025 Dec;28(1):766-777. doi: 10.1080/13696998.2025.2503660. Epub 2025 May 23. J Med Econ. 2025. PMID: 40395149
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Olaparib Monotherapy or in Combination with Abiraterone for the Treatment of Patients with Metastatic Castration-Resistant Prostate Cancer (mCRPC) and a BRCA Mutation.Target Oncol. 2025 May;20(3):445-466. doi: 10.1007/s11523-025-01146-4. Epub 2025 May 21. Target Oncol. 2025. PMID: 40397306 Free PMC article. Review.
Cited by
-
Elevated VRK1 levels after androgen deprivation therapy promote prostate cancer progression by upregulating YAP1 expression.J Cancer Res Clin Oncol. 2025 Mar 20;151(3):116. doi: 10.1007/s00432-025-06168-z. J Cancer Res Clin Oncol. 2025. PMID: 40111564 Free PMC article.
-
RIOK1: A Novel Oncogenic Driver in Hepatocellular Carcinoma.Cancer Med. 2025 Feb;14(3):e70597. doi: 10.1002/cam4.70597. Cancer Med. 2025. PMID: 39865406 Free PMC article.
-
MED12 and CDK8/19 Modulate Androgen Receptor Activity and Enzalutamide Response in Prostate Cancer.Endocrinology. 2024 Aug 27;165(10):bqae114. doi: 10.1210/endocr/bqae114. Endocrinology. 2024. PMID: 39253786 Free PMC article.
-
The RioK1 network determines p53 activity at multiple levels.Cell Death Discov. 2023 Nov 7;9(1):410. doi: 10.1038/s41420-023-01704-7. Cell Death Discov. 2023. PMID: 37935656 Free PMC article.
References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Mottet N., van den Bergh R.C.N., Briers E., Van den Broeck T., Cumberbatch M.G., De Santis M., Fanti S., Fossati N., Gandaglia G., Gillessen S., Grivas N., Grummet J., Henry A.M., van der Kwast T.H., Lam T.B., Lardas M., Liew M., Mason M.D., Moris L., Oprea-Lager D.E., van der Poel H.G., Rouviere O., Schoots I.G., Tilki D., Wiegel T., Willemse P.M., Cornford P. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer-2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2021;79:243–262. - PubMed
-
- Kanev G.K., de Graaf C., de Esch I.J.P., Leurs R., Wurdinger T., Westerman B.A., Kooistra A.J. The landscape of atypical and eukaryotic protein kinases. Trends Pharmacol Sci. 2019;40:818–832. - PubMed
-
- Chau V., Madan R.A., Aragon-Ching J.B. Protein kinase inhibitors for the treatment of prostate cancer. Expert Opin Pharmacother. 2021;22:1889–1899. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

