Plasma proteome of Long-COVID patients indicates HIF-mediated vasculo-proliferative disease with impact on brain and heart function
- PMID: 37301958
- PMCID: PMC10257382
- DOI: 10.1186/s12967-023-04149-9
Plasma proteome of Long-COVID patients indicates HIF-mediated vasculo-proliferative disease with impact on brain and heart function
Abstract
Aims: Long-COVID occurs after SARS-CoV-2 infection and results in diverse, prolonged symptoms. The present study aimed to unveil potential mechanisms, and to inform prognosis and treatment.
Methods: Plasma proteome from Long-COVID outpatients was analyzed in comparison to matched acutely ill COVID-19 (mild and severe) inpatients and healthy control subjects. The expression of 3072 protein biomarkers was determined with proximity extension assays and then deconvoluted with multiple bioinformatics tools into both cell types and signaling mechanisms, as well as organ specificity.
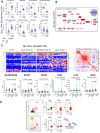
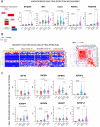
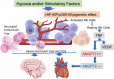
Results: Compared to age- and sex-matched acutely ill COVID-19 inpatients and healthy control subjects, Long-COVID outpatients showed natural killer cell redistribution with a dominant resting phenotype, as opposed to active, and neutrophils that formed extracellular traps. This potential resetting of cell phenotypes was reflected in prospective vascular events mediated by both angiopoietin-1 (ANGPT1) and vascular-endothelial growth factor-A (VEGFA). Several markers (ANGPT1, VEGFA, CCR7, CD56, citrullinated histone 3, elastase) were validated by serological methods in additional patient cohorts. Signaling of transforming growth factor-β1 with probable connections to elevated EP/p300 suggested vascular inflammation and tumor necrosis factor-α driven pathways. In addition, a vascular proliferative state associated with hypoxia inducible factor 1 pathway suggested progression from acute COVID-19 to Long-COVID. The vasculo-proliferative process predicted in Long-COVID might contribute to changes in the organ-specific proteome reflective of neurologic and cardiometabolic dysfunction.
Conclusions: Taken together, our findings point to a vasculo-proliferative process in Long-COVID that is likely initiated either prior hypoxia (localized or systemic) and/or stimulatory factors (i.e., cytokines, chemokines, growth factors, angiotensin, etc). Analyses of the plasma proteome, used as a surrogate for cellular signaling, unveiled potential organ-specific prognostic biomarkers and therapeutic targets.
© 2023. The Author(s).
Conflict of interest statement
There are no competing interests.
Figures






References
-
- World Health Organization WHO R&D Blueprint. Novel Coronavirus. COVID-19 Therapeutic Trial Synopsis.https://cdn.who.int/media/docs/default-source/blue-print/covid-19-therap....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

