Staphylococcus epidermidis isolates from atopic or healthy skin have opposite effect on skin cells: potential implication of the AHR pathway modulation
- PMID: 37304256
- PMCID: PMC10250813
- DOI: 10.3389/fimmu.2023.1098160
Staphylococcus epidermidis isolates from atopic or healthy skin have opposite effect on skin cells: potential implication of the AHR pathway modulation
Abstract
Introduction: Staphylococcus epidermidis is a commensal bacterium ubiquitously present on human skin. This species is considered as a key member of the healthy skin microbiota, involved in the defense against pathogens, modulating the immune system, and involved in wound repair. Simultaneously, S. epidermidis is the second cause of nosocomial infections and an overgrowth of S. epidermidis has been described in skin disorders such as atopic dermatitis. Diverse isolates of S. epidermidis co-exist on the skin. Elucidating the genetic and phenotypic specificities of these species in skin health and disease is key to better understand their role in various skin conditions. Additionally, the exact mechanisms by which commensals interact with host cells is partially understood. We hypothesized that S. epidermidis isolates identified from different skin origins could play distinct roles on skin differentiation and that these effects could be mediated by the aryl hydrocarbon receptor (AhR) pathway.
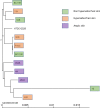
Methods: For this purpose, a library of 12 strains originated from healthy skin (non-hyperseborrheic (NH) and hyperseborrheic (H) skin types) and disease skin (atopic (AD) skin type) was characterized at the genomic and phenotypic levels.
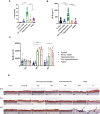
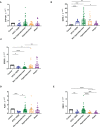
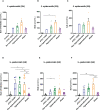
Results and discussion: Here we showed that strains from atopic lesional skin alter the epidermis structure of a 3D reconstructed skin model whereas strains from NH healthy skin do not. All strains from NH healthy skin induced AhR/OVOL1 path and produced high quantities of indole metabolites in co-culture with NHEK; especially indole-3-aldehyde (IAld) and indole-3-lactic acid (ILA); while AD strains did not induce AhR/OVOL1 path but its inhibitor STAT6 and produced the lowest levels of indoles as compared to the other strains. As a consequence, strains from AD skin altered the differentiation markers FLG and DSG1. The results presented here, on a library of 12 strains, showed that S. epidermidis originated from NH healthy skin and atopic skin have opposite effects on the epidermal cohesion and structure and that these differences could be linked to their capacity to produce metabolites, which in turn could activate AHR pathway. Our results on a specific library of strains provide new insights into how S. epidermidis may interact with the skin to promote health or disease.
Keywords: S. epidermidis; aryl hydrocarbon receptor; biofilms; indoles; inflammation; proteases; skin microbiome; skin type.
Copyright © 2023 Landemaine, Da Costa, Fissier, Francis, Morand, Verbeke, Michel, Briandet, Sokol, Gueniche, Bernard, Chatel, Aguilar, Langella, Clavaud and Richard.
Conflict of interest statement
Authors LL, CC, CF, SM, AG, DB and LA were employed by the company L’Oréal Research and Innovation, Aulnay-sous-Bois. JV was employed by iMEAN, Toulouse, France. L’Oréal Research and Innovation, as funder, has been involved in the decision to submit the study for publication.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

