Analyses of the Dmrt family in a decapod crab, Eriocheir sinensis uncover new facets on the evolution of DM domain genes
- PMID: 37304820
- PMCID: PMC10252143
- DOI: 10.3389/fphys.2023.1201846
Analyses of the Dmrt family in a decapod crab, Eriocheir sinensis uncover new facets on the evolution of DM domain genes
Abstract
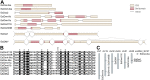
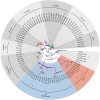
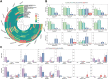
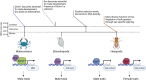
DM domain genes are a group of transcription factors that are integral to sexual development and its evolution in metazoans. Their functions and regulatory mechanisms are not well understood in Malacostraca (crabs and crayfish) while these sex regulators have been widely identified in the past decade. In this study, the Dmrt family was investigated in the decapod crab, Eriocheir sinensis. We find that most members of the EsDmrt family begin to enrich around the juvenile 1 stage. In reproductive organs, EsDsx1, EsDsx2, EsiDMY and EsiDmrt1a highly express in the male-specific androgenic gland (AG), while EsDmrt-like, EsDsx-like, EsDmrt11E, and EsiDmrt1b show relatively high expression in testis. Also, we find the highly aberrant expression of EsiDMY and EsiDmrt1a in the chimeric AG, strongly indicating their function in AG development. Moreover, RNA interference of EsDsx1, EsiDMY, and EsiDmrt1a results in a significant decrease in transcription of the Insulin-like androgenic hormone (IAG), respectively. Our findings suggest that Dmrt genes in E. sinensis primarily function in male sexual differentiation, especially in AG development. Besides, this study identifies two unique groups of Dmrt genes in Malacostraca: Dsx and iDmrt1. In Malacostraca Dsx, we uncover a cryptic mutation in the eight zinc motif-specific residues, which were firmly believed to be invariant across the Dmrt family. This mutation sets the Malacostraca Dsx apart from all the other Dmrt genes and implies a different way of transcriptional regulation. Genes from the iDmrt1 group show phylogenetical limitation to the malacostracan species and underwent positive selection, suggesting their highly specialized gene function to this class. Based on these findings, we propose that Dsx and iDmrt1 in Malacostraca have developed unique transcriptional regulation mechanisms to facilitate AG development. We hope that this study would contribute to our understandings of sexual development in Malacostraca and provide new insights into the evolutionary history of the Dmrt family.
Keywords: Dmrt; Eriocheir sinensis; evolution; malacostraca; sexual development.
Copyright © 2023 Zhang, Yang, Xu and Cui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis.Dev Genes Evol. 2010 Nov;220(5-6):151-9. doi: 10.1007/s00427-010-0336-2. Epub 2010 Aug 31. Dev Genes Evol. 2010. PMID: 20809137
-
miR-34 negatively regulates the expression of Dmrt and related genes in the testis of mud crab Scylla paramamosain.Comp Biochem Physiol B Biochem Mol Biol. 2025 Jan;275:111018. doi: 10.1016/j.cbpb.2024.111018. Epub 2024 Aug 14. Comp Biochem Physiol B Biochem Mol Biol. 2025. PMID: 39128537
-
Identification and functional analysis of the doublesex gene in the mud crab Scylla paramamosain.Comp Biochem Physiol A Mol Integr Physiol. 2022 Apr;266:111150. doi: 10.1016/j.cbpa.2022.111150. Epub 2022 Jan 10. Comp Biochem Physiol A Mol Integr Physiol. 2022. PMID: 35017065
-
The "IAG-Switch"-A Key Controlling Element in Decapod Crustacean Sex Differentiation.Front Endocrinol (Lausanne). 2020 Sep 10;11:651. doi: 10.3389/fendo.2020.00651. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33013714 Free PMC article. Review.
-
Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis.Cell Mol Life Sci. 2013 Oct;70(20):3829-45. doi: 10.1007/s00018-013-1288-2. Epub 2013 Mar 5. Cell Mol Life Sci. 2013. PMID: 23463235 Free PMC article. Review.
Cited by
-
DNA Methylation Patterns Provide Insights into the Epigenetic Regulation of Intersex Formation in the Chinese Mitten Crab (Eriocheir sinensis).Int J Mol Sci. 2025 Mar 30;26(7):3224. doi: 10.3390/ijms26073224. Int J Mol Sci. 2025. PMID: 40244073 Free PMC article.
-
Monosex Populations of the Giant Freshwater Prawn Macrobrachium rosenbergii-From a Pre-Molecular Start to the Next Generation Era.Int J Mol Sci. 2023 Dec 13;24(24):17433. doi: 10.3390/ijms242417433. Int J Mol Sci. 2023. PMID: 38139271 Free PMC article. Review.
References
-
- Chandler J. C., Elizur A., Ventura T. (2018). The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 825, 61–80. 10.1007/s10750-017-3452-4 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

