Combining multiplex gene editing and doubled haploid technology in maize
- PMID: 37306056
- PMCID: PMC7614789
- DOI: 10.1111/nph.19021
Combining multiplex gene editing and doubled haploid technology in maize
Abstract
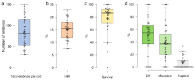
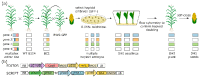
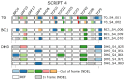
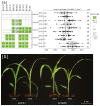
A major advantage of using CRISPR/Cas9 for gene editing is multiplexing, that is, the simultaneous targeting of many genes. However, primary transformants typically contain hetero-allelic mutations or are genetic mosaic, while genetically stable lines that are homozygous are desired for functional analysis. Currently, a dedicated and labor-intensive effort is required to obtain such higher-order mutants through several generations of genetic crosses and genotyping. We describe the design and validation of a rapid and efficient strategy to produce lines of genetically identical plants carrying various combinations of homozygous edits, suitable for replicated analysis of phenotypical differences. This approach was achieved by combining highly multiplex gene editing in Zea mays (maize) with in vivo haploid induction and efficient in vitro generation of doubled haploid plants using embryo rescue doubling. By combining three CRISPR/Cas9 constructs that target in total 36 genes potentially involved in leaf growth, we generated an array of homozygous lines with various combinations of edits within three generations. Several genotypes show a reproducible 10% increase in leaf size, including a septuple mutant combination. We anticipate that our strategy will facilitate the study of gene families via multiplex CRISPR mutagenesis and the identification of allele combinations to improve quantitative crop traits.
Keywords: CRISPR/Cas9; doubled haploids; gene editing; gene family; haploid induction; maize; multiplex gene editing; mutation stacking.
© 2023 The Authors. New Phytologist © 2023 New Phytologist Foundation.
Conflict of interest statement
All authors declare no competing interests.
Figures


Similar articles
-
Development of a Haploid-Inducer Mediated Genome Editing System for Accelerating Maize Breeding.Mol Plant. 2019 Apr 1;12(4):597-602. doi: 10.1016/j.molp.2019.03.006. Epub 2019 Mar 19. Mol Plant. 2019. PMID: 30902686
-
Harnessing haploid-inducer mediated genome editing for accelerated maize variety development.Plant Biotechnol J. 2025 May;23(5):1604-1614. doi: 10.1111/pbi.14608. Epub 2025 Feb 12. Plant Biotechnol J. 2025. PMID: 39936495 Free PMC article.
-
CRISPR/Cas9-Mediated Targeted Mutagenesis in Wheat Doubled Haploids.Methods Mol Biol. 2020;2072:183-198. doi: 10.1007/978-1-4939-9865-4_15. Methods Mol Biol. 2020. PMID: 31541447
-
Advances in Gene Editing of Haploid Tissues in Crops.Genes (Basel). 2021 Sep 13;12(9):1410. doi: 10.3390/genes12091410. Genes (Basel). 2021. PMID: 34573392 Free PMC article. Review.
-
Maize In Planta Haploid Inducer Lines: A Cornerstone for Doubled Haploid Technology.Methods Mol Biol. 2021;2288:25-48. doi: 10.1007/978-1-0716-1335-1_2. Methods Mol Biol. 2021. PMID: 34270003 Review.
Cited by
-
Breeding for improved digestibility and processing of lignocellulosic biomass in Zea mays.Front Plant Sci. 2024 Jul 26;15:1419796. doi: 10.3389/fpls.2024.1419796. eCollection 2024. Front Plant Sci. 2024. PMID: 39129761 Free PMC article. Review.
-
Green revolution to genome revolution: driving better resilient crops against environmental instability.Front Genet. 2023 Aug 31;14:1204585. doi: 10.3389/fgene.2023.1204585. eCollection 2023. Front Genet. 2023. PMID: 37719711 Free PMC article. Review.
-
Unlocking Nature's Clock: CRISPR Technology in Flowering Time Engineering.Plants (Basel). 2023 Nov 29;12(23):4020. doi: 10.3390/plants12234020. Plants (Basel). 2023. PMID: 38068655 Free PMC article. Review.
-
Genetic Analysis and Construction of a Fingerprint for Licensed Triadica sebifera Cultivars Using SSR Markers.Plants (Basel). 2024 Jun 26;13(13):1767. doi: 10.3390/plants13131767. Plants (Basel). 2024. PMID: 38999607 Free PMC article.
-
Roles of auxin pathways in maize biology.J Exp Bot. 2023 Dec 1;74(22):6989-6999. doi: 10.1093/jxb/erad297. J Exp Bot. 2023. PMID: 37493143 Free PMC article. Review.
References
-
- Aesaert S, Impens L, Coussens G, Van Lerberge E, Vanderhaeghen R, Desmet L, Vanhevel Y, Bossuyt S, Wambua AN, Van Lijsebettens M, et al. Optimized transformation and gene editing of the B104 public maize inbred by improved tissue culture and use of morphogenic regulators. Frontiers in Plant Science. 2022;13:883847. - PMC - PubMed
-
- Barton JE, Maddock SE, Wu XE, Zhao Z-Y, Williams ME, Hussain T, Gordon-Kamm WJ. Doubling of chromosomes in haploid embryos. 2014. https://patents.google.com/patent/US8859846B2/en .
-
- Bonnett D, Rebetzke G, Spielmeyer W. Strategies for efficient implementation of molecular markers in wheat breeding. Molecular Breeding. 2005;15:75–85.
-
- Bregitzer P, Dahleen LS, Neate S, Schwarz P, Manoharan M. A single backcross effectively eliminates agronomic and quality alterations caused by somaclonal variation in transgenic barley. Crop Science. 2008;48:471–479.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

