A Novel Conserved Linear Neutralizing Epitope on the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein
- PMID: 37306579
- PMCID: PMC10433833
- DOI: 10.1128/spectrum.01190-23
A Novel Conserved Linear Neutralizing Epitope on the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein
Abstract
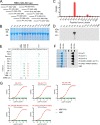
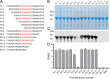
The continuous emergence of new variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has made it challenging to develop broad-spectrum prophylactic vaccines and therapeutic antibodies. Here, we have identified a broad-spectrum neutralizing antibody and its highly conserved epitope in the receptor-binding domain (RBD) of the spike protein (S) S1 subunit of SARS-CoV-2. First, nine monoclonal antibodies (MAbs) against the RBD or S1 were generated; of these, one RBD-specific MAb, 22.9-1, was selected for its broad RBD-binding abilities and neutralizing activities against SARS-CoV-2 variants. An epitope of 22.9-1 was fine-mapped with overlapping and truncated peptide fusion proteins. The core sequence of the epitope, 405D(N)EVR(S)QIAPGQ414, was identified on the internal surface of the up-state RBD. The epitope was conserved in nearly all variants of concern of SARS-CoV-2. MAb 22.9-1 and its novel epitope could be beneficial for research on broad-spectrum prophylactic vaccines and therapeutic antibody drugs. IMPORTANCE The continuous emergence of new variants of SARS-CoV-2 has caused great challenge in vaccine design and therapeutic antibody development. In this study, we selected a broad-spectrum neutralizing mouse monoclonal antibody which recognized a conserved linear B-cell epitope located on the internal surface of RBD. This MAb could neutralize all variants until now. The epitope was conserved in all variants. This work provides new insights in developing broad-spectrum prophylactic vaccines and therapeutic antibodies.
Keywords: SARS-CoV-2; epitope; neutralizing antibody; receptor-binding domain; spike protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Structural Basis of a Human Neutralizing Antibody Specific to the SARS-CoV-2 Spike Protein Receptor-Binding Domain.Microbiol Spectr. 2021 Oct 31;9(2):e0135221. doi: 10.1128/Spectrum.01352-21. Epub 2021 Oct 13. Microbiol Spectr. 2021. PMID: 34643438 Free PMC article.
-
A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection.J Virol. 2022 Sep 14;96(17):e0011822. doi: 10.1128/jvi.00118-22. Epub 2022 Aug 16. J Virol. 2022. PMID: 35972290 Free PMC article.
-
Characterization of a neutralizing antibody that recognizes a loop region adjacent to the receptor-binding interface of the SARS-CoV-2 spike receptor-binding domain.Microbiol Spectr. 2024 Apr 2;12(4):e0365523. doi: 10.1128/spectrum.03655-23. Epub 2024 Feb 28. Microbiol Spectr. 2024. PMID: 38415660 Free PMC article.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting.Int J Mol Sci. 2022 Apr 14;23(8):4341. doi: 10.3390/ijms23084341. Int J Mol Sci. 2022. PMID: 35457159 Free PMC article. Review.
Cited by
-
Identification of Two Linear Epitopes on MGF_110-13L Protein of African Swine Fever Virus with Monoclonal Antibodies.Animals (Basel). 2024 Jul 1;14(13):1951. doi: 10.3390/ani14131951. Animals (Basel). 2024. PMID: 38998063 Free PMC article.
-
A novel monospecific tetravalent IgG1-(scFv)2 version shown enhanced neutralizing and Fc-mediated effector functions against SARS-CoV-2.3 Biotech. 2023 Aug;13(8):283. doi: 10.1007/s13205-023-03702-z. Epub 2023 Jul 25. 3 Biotech. 2023. PMID: 37501919 Free PMC article.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP, Bjorkman PJ. 2020. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588:682–687. doi:10.1038/s41586-020-2852-1. - DOI - PMC - PubMed
-
- Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann H-H, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9:e61312. doi:10.7554/eLife.61312. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

