Landscape of New Nuclease-Containing Antiphage Systems in Escherichia coli and the Counterdefense Roles of Bacteriophage T4 Genome Modifications
- PMID: 37306585
- PMCID: PMC10308915
- DOI: 10.1128/jvi.00599-23
Landscape of New Nuclease-Containing Antiphage Systems in Escherichia coli and the Counterdefense Roles of Bacteriophage T4 Genome Modifications
Abstract
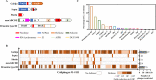
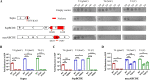
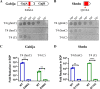
Many phages, such as T4, protect their genomes against the nucleases of bacterial restriction-modification (R-M) and CRISPR-Cas systems through covalent modification of their genomes. Recent studies have revealed many novel nuclease-containing antiphage systems, raising the question of the role of phage genome modifications in countering these systems. Here, by focusing on phage T4 and its host Escherichia coli, we depicted the landscape of the new nuclease-containing systems in E. coli and demonstrated the roles of T4 genome modifications in countering these systems. Our analysis identified at least 17 nuclease-containing defense systems in E. coli, with type III Druantia being the most abundant system, followed by Zorya, Septu, Gabija, AVAST type 4, and qatABCD. Of these, 8 nuclease-containing systems were found to be active against phage T4 infection. During T4 replication in E. coli, 5-hydroxymethyl dCTP is incorporated into the newly synthesized DNA instead of dCTP. The 5-hydroxymethylcytosines (hmCs) are further modified by glycosylation to form glucosyl-5-hydroxymethylcytosine (ghmC). Our data showed that the ghmC modification of the T4 genome abolished the defense activities of Gabija, Shedu, Restriction-like, type III Druantia, and qatABCD systems. The anti-phage T4 activities of the last two systems can also be counteracted by hmC modification. Interestingly, the Restriction-like system specifically restricts phage T4 containing an hmC-modified genome. The ghmC modification cannot abolish the anti-phage T4 activities of Septu, SspBCDE, and mzaABCDE, although it reduces their efficiency. Our study reveals the multidimensional defense strategies of E. coli nuclease-containing systems and the complex roles of T4 genomic modification in countering these defense systems. IMPORTANCE Cleavage of foreign DNA is a well-known mechanism used by bacteria to protect themselves from phage infections. Two well-known bacterial defense systems, R-M and CRISPR-Cas, both contain nucleases that cleave the phage genomes through specific mechanisms. However, phages have evolved different strategies to modify their genomes to prevent cleavage. Recent studies have revealed many novel nuclease-containing antiphage systems from various bacteria and archaea. However, no studies have systematically investigated the nuclease-containing antiphage systems of a specific bacterial species. In addition, the role of phage genome modifications in countering these systems remains unknown. Here, by focusing on phage T4 and its host Escherichia coli, we depicted the landscape of the new nuclease-containing systems in E. coli using all 2,289 genomes available in NCBI. Our studies reveal the multidimensional defense strategies of E. coli nuclease-containing systems and the complex roles of genomic modification of phage T4 in countering these defense systems.
Keywords: E. coli defense systems; bacteriophage; genome modifications; phage T4.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Covalent Modifications of the Bacteriophage Genome Confer a Degree of Resistance to Bacterial CRISPR Systems.J Virol. 2020 Nov 9;94(23):e01630-20. doi: 10.1128/JVI.01630-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32938767 Free PMC article.
-
Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9.mBio. 2015 Jun 16;6(3):e00648. doi: 10.1128/mBio.00648-15. mBio. 2015. PMID: 26081634 Free PMC article.
-
Bacteriophage T4 Escapes CRISPR Attack by Minihomology Recombination and Repair.mBio. 2021 Jun 29;12(3):e0136121. doi: 10.1128/mBio.01361-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154416 Free PMC article.
-
Anticodon nucleases.Trends Biochem Sci. 2000 Feb;25(2):70-4. doi: 10.1016/s0968-0004(99)01525-x. Trends Biochem Sci. 2000. PMID: 10664586 Review.
-
OLD family nuclease function across diverse anti-phage defense systems.Front Microbiol. 2023 Sep 28;14:1268820. doi: 10.3389/fmicb.2023.1268820. eCollection 2023. Front Microbiol. 2023. PMID: 37840731 Free PMC article. Review.
Cited by
-
Correlation of Pseudomonas aeruginosa Phage Resistance with the Numbers and Types of Antiphage Systems.Int J Mol Sci. 2024 Jan 24;25(3):1424. doi: 10.3390/ijms25031424. Int J Mol Sci. 2024. PMID: 38338703 Free PMC article.
-
A Class 1 OLD family nuclease encoded by Vibrio cholerae is countered by a vibriophage-encoded direct inhibitor.bioRxiv [Preprint]. 2025 Jan 6:2025.01.06.631583. doi: 10.1101/2025.01.06.631583. bioRxiv. 2025. PMID: 39829814 Free PMC article. Preprint.
-
Bacterial Hachiman complex executes DNA cleavage for antiphage defense.Nat Commun. 2025 Mar 17;16(1):2604. doi: 10.1038/s41467-025-57851-1. Nat Commun. 2025. PMID: 40097437 Free PMC article.
-
Structural Basis for Retron Co-option of Anti-phage ATPase-nuclease.bioRxiv [Preprint]. 2025 May 13:2025.05.10.653283. doi: 10.1101/2025.05.10.653283. bioRxiv. 2025. PMID: 40462907 Free PMC article. Preprint.
-
PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.Nat Struct Mol Biol. 2024 Mar;31(3):413-423. doi: 10.1038/s41594-023-01172-8. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177683 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

