MBD2a-NuRD binds to the methylated γ-globin gene promoter and uniquely forms a complex required for silencing of HbF expression
- PMID: 37307480
- PMCID: PMC10288633
- DOI: 10.1073/pnas.2302254120
MBD2a-NuRD binds to the methylated γ-globin gene promoter and uniquely forms a complex required for silencing of HbF expression
Abstract
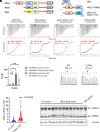
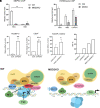
During human development, there is a switch in the erythroid compartment at birth that results in silencing of expression of fetal hemoglobin (HbF). Reversal of this silencing has been shown to be effective in overcoming the pathophysiologic defect in sickle cell anemia. Among the many transcription factors and epigenetic effectors that are known to mediate HbF silencing, two of the most potent are BCL11A and MBD2-NuRD. In this report, we present direct evidence that MBD2-NuRD occupies the γ-globin gene promoter in adult erythroid cells and positions a nucleosome there that results in a closed chromatin conformation that prevents binding of the transcriptional activator, NF-Y. We show that the specific isoform, MBD2a, is required for the formation and stable occupancy of this repressor complex that includes BCL11A, MBD2a-NuRD, and the arginine methyltransferase, PRMT5. The methyl cytosine binding preference and the arginine-rich (GR) domain of MBD2a are required for high affinity binding to methylated γ-globin gene proximal promoter DNA sequences. Mutation of the methyl cytosine-binding domain (MBD) of MBD2 results in a variable but consistent loss of γ-globin gene silencing, in support of the importance of promoter methylation. The GR domain of MBD2a is also required for recruitment of PRMT5, which in turn results in placement of the repressive chromatin mark H3K8me2s at the promoter. These findings support a unified model that integrates the respective roles of BCL11A, MBD2a-NuRD, PRMT5, and DNA methylation in HbF silencing.
Keywords: BCL11A; DNA methylation; MBD2a–NuRD; PRMT5; fetal hemoglobin.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Sankaran V. G., et al. , Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322, 1839–1842 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

