Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis
- PMID: 37308477
- PMCID: PMC10261145
- DOI: 10.1038/s41467-023-39274-y
Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis
Abstract
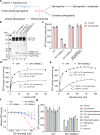
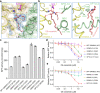
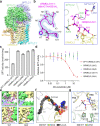
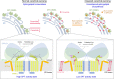
The ORM/ORMDL family proteins function as regulatory subunits of the serine palmitoyltransferase (SPT) complex, which is the initiating and rate-limiting enzyme in sphingolipid biosynthesis. This complex is tightly regulated by cellular sphingolipid levels, but the sphingolipid sensing mechanism is unknown. Here we show that purified human SPT-ORMDL complexes are inhibited by the central sphingolipid metabolite ceramide. We have solved the cryo-EM structure of the SPT-ORMDL3 complex in a ceramide-bound state. Structure-guided mutational analyses reveal the essential function of this ceramide binding site for the suppression of SPT activity. Structural studies indicate that ceramide can induce and lock the N-terminus of ORMDL3 into an inhibitory conformation. Furthermore, we demonstrate that childhood amyotrophic lateral sclerosis (ALS) variants in the SPTLC1 subunit cause impaired ceramide sensing in the SPT-ORMDL3 mutants. Our work elucidates the molecular basis of ceramide sensing by the SPT-ORMDL complex for establishing sphingolipid homeostasis and indicates an important role of impaired ceramide sensing in disease development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

