Single cell and spatial sequencing define processes by which keratinocytes and fibroblasts amplify inflammatory responses in psoriasis
- PMID: 37308489
- PMCID: PMC10261041
- DOI: 10.1038/s41467-023-39020-4
Single cell and spatial sequencing define processes by which keratinocytes and fibroblasts amplify inflammatory responses in psoriasis
Abstract
The immunopathogenesis of psoriasis, a common chronic inflammatory disease of the skin, is incompletely understood. Here we demonstrate, using a combination of single cell and spatial RNA sequencing, IL-36 dependent amplification of IL-17A and TNF inflammatory responses in the absence of neutrophil proteases, which primarily occur within the supraspinous layer of the psoriatic epidermis. We further show that a subset of SFRP2+ fibroblasts in psoriasis contribute to amplification of the immune network through transition to a pro-inflammatory state. The SFRP2+ fibroblast communication network involves production of CCL13, CCL19 and CXCL12, connected by ligand-receptor interactions to other spatially proximate cell types: CCR2+ myeloid cells, CCR7+ LAMP3+ dendritic cells, and CXCR4 expressed on both CD8+ Tc17 cells and keratinocytes, respectively. The SFRP2+ fibroblasts also express cathepsin S, further amplifying inflammatory responses by activating IL-36G in keratinocytes. These data provide an in-depth view of psoriasis pathogenesis, which expands our understanding of the critical cellular participants to include inflammatory fibroblasts and their cellular interactions.
© 2023. The Author(s).
Conflict of interest statement
J.E.G. has received research support from Almirall, AnaptysBio, Novartis, AbbVie, Eli Lilly, Kyowa Kirin, Galderma, BMS, Janssen. J.E.G. has served as advisor to Pfizer, AbbVie, Eli Lilly, Galderma, Almirall, AnaptysBio, Novartis, Sanofi, BMS. J.M.K. has received research support from Janssen, BMS, and Q32 bio. J.M.K. has served as an advisor to Eli Lilly, BMS, AstraZeneca, GlaxoSmithKline, Ventus Therapeutics, Admirex Pharmaceuticals, Aurinia Pharmaceuticals, and Provention Bio. The remaining authors declare no competing interests.
Figures
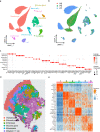



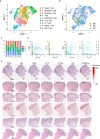

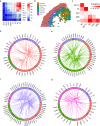
References
Publication types
MeSH terms
Grants and funding
- R01 AR073196/AR/NIAMS NIH HHS/United States
- R01 AR075959/AR/NIAMS NIH HHS/United States
- R01 AR042742/AR/NIAMS NIH HHS/United States
- R01 AR054966/AR/NIAMS NIH HHS/United States
- UG3 CA268091/CA/NCI NIH HHS/United States
- UC2 AR081029/AR/NIAMS NIH HHS/United States
- R25 GM086262/GM/NIGMS NIH HHS/United States
- P50 AR070590/AR/NIAMS NIH HHS/United States
- R01 AR040312/AR/NIAMS NIH HHS/United States
- R01 AR069071/AR/NIAMS NIH HHS/United States
- R01 AI022553/AI/NIAID NIH HHS/United States
- R01 AR074302/AR/NIAMS NIH HHS/United States
- K08 AR078251/AR/NIAMS NIH HHS/United States
- P30 AR075043/AR/NIAMS NIH HHS/United States
- K01 AR072129/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

