A rapid and bidirectional reporter of neural activity reveals neural correlates of social behaviors in Drosophila
- PMID: 37308660
- PMCID: PMC10866131
- DOI: 10.1038/s41593-023-01357-w
A rapid and bidirectional reporter of neural activity reveals neural correlates of social behaviors in Drosophila
Abstract
Neural activity is modulated over different timescales encompassing subseconds to hours, reflecting changes in external environment, internal state and behavior. Using Drosophila as a model, we developed a rapid and bidirectional reporter that provides a cellular readout of recent neural activity. This reporter uses nuclear versus cytoplasmic distribution of CREB-regulated transcriptional co-activator (CRTC). Subcellular distribution of GFP-tagged CRTC (CRTC::GFP) bidirectionally changes on the order of minutes and reflects both increases and decreases in neural activity. We established an automated machine-learning-based routine for efficient quantification of reporter signal. Using this reporter, we demonstrate mating-evoked activation and inactivation of modulatory neurons. We further investigated the functional role of the master courtship regulator gene fruitless (fru) and show that fru is necessary to ensure activation of male arousal neurons by female cues. Together, our results establish CRTC::GFP as a bidirectional reporter of recent neural activity suitable for examining neural correlates in behavioral contexts.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




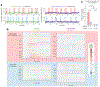
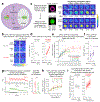
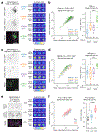
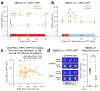


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

