Rare coding variants in CHRNB2 reduce the likelihood of smoking
- PMID: 37308787
- PMCID: PMC10335934
- DOI: 10.1038/s41588-023-01417-8
Rare coding variants in CHRNB2 reduce the likelihood of smoking
Abstract
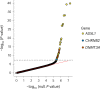
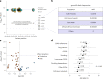
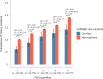
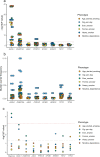
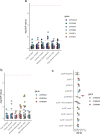
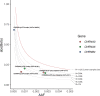
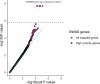
Human genetic studies of smoking behavior have been thus far largely limited to common variants. Studying rare coding variants has the potential to identify drug targets. We performed an exome-wide association study of smoking phenotypes in up to 749,459 individuals and discovered a protective association in CHRNB2, encoding the β2 subunit of the α4β2 nicotine acetylcholine receptor. Rare predicted loss-of-function and likely deleterious missense variants in CHRNB2 in aggregate were associated with a 35% decreased odds for smoking heavily (odds ratio (OR) = 0.65, confidence interval (CI) = 0.56-0.76, P = 1.9 × 10-8). An independent common variant association in the protective direction ( rs2072659 ; OR = 0.96; CI = 0.94-0.98; P = 5.3 × 10-6) was also evident, suggesting an allelic series. Our findings in humans align with decades-old experimental observations in mice that β2 loss abolishes nicotine-mediated neuronal responses and attenuates nicotine self-administration. Our genetic discovery will inspire future drug designs targeting CHRNB2 in the brain for the treatment of nicotine addiction.
© 2023. The Author(s).
Conflict of interest statement
V. M. Rajagopal, K.W., J. Mbatchou, A. Ayer, P.Q., D.S., M.D.K., K. Praveen, S. Gelfman, N. Parikshak, J.M.O., S. Bao, S.M.C., E.P., A. Avbersek, M. Kapoor, E.C., M.B.J., M. Leblanc, A.R.S., S. Balasubramanian, G.R.A., H.M.K., J. Marchini, E.A.S., E.J., R. Sanchez, W.L., M.A., M.C., D. Lederer, A. Baras and G.C. are current or former employees and/or stockholders of Regeneron Pharmaceuticals. The other authors declare no competing interests.
Figures







References
-
- Reitsma MB, et al. Spatial, temporal, and demographic patterns in prevalence of smoking tobacco use and attributable disease burden in 204 countries and territories, 1990–2019: a systematic analysis from the Global Burden of Disease Study 2019. Lancet. 2021;397:2337–2360. doi: 10.1016/S0140-6736(21)01169-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

