Systematic approaches to C-lignin engineering in Medicago truncatula
- PMID: 37308891
- PMCID: PMC10262568
- DOI: 10.1186/s13068-023-02339-7
Systematic approaches to C-lignin engineering in Medicago truncatula
Abstract
Background: C-lignin is a homopolymer of caffeyl alcohol present in the seed coats of a variety of plant species including vanilla orchid, various cacti, and the ornamental plant Cleome hassleriana. Because of its unique chemical and physical properties, there is considerable interest in engineering C-lignin into the cell walls of bioenergy crops as a high-value co-product of bioprocessing. We have used information from a transcriptomic analysis of developing C. hassleriana seed coats to suggest strategies for engineering C-lignin in a heterologous system, using hairy roots of the model legume Medicago truncatula.
Results: We systematically tested strategies for C-lignin engineering using a combination of gene overexpression and RNAi-mediated knockdown in the caffeic acid/5-hydroxy coniferaldehyde 3/5-O-methyltransferase (comt) mutant background, monitoring the outcomes by analysis of lignin composition and profiling of monolignol pathway metabolites. In all cases, C-lignin accumulation required strong down-regulation of caffeoyl CoA 3-O-methyltransferase (CCoAOMT) paired with loss of function of COMT. Overexpression of the Selaginella moellendorffii ferulate 5-hydroxylase (SmF5H) gene in comt mutant hairy roots resulted in lines that unexpectedly accumulated high levels of S-lignin.
Conclusion: C-Lignin accumulation of up to 15% of total lignin in lines with the greatest reduction in CCoAOMT expression required the strong down-regulation of both COMT and CCoAOMT, but did not require expression of a heterologous laccase, cinnamyl alcohol dehydrogenase (CAD) or cinnamoyl CoA reductase (CCR) with preference for 3,4-dihydroxy-substituted substrates in M. truncatula hairy roots. Cell wall fractionation studies suggested that the engineered C-units are not present in a heteropolymer with the bulk of the G-lignin.
Keywords: C-lignin; Co-product; Hairy roots; Medicago truncatula; Metabolic engineering; Transgenic plants.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

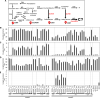
 sign. The 3-OH group in caffeyl alcohol can be introduced by the direct action of C3H on coumarate, or by the combined activities of HCT, C3′H and CSE in the so-called “shikimate shunt”. S lignin is not made in the Cleome seed coat due to lack of expression of F5H. Enzymes are L-phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT), coumaroyl shikimate 3′-hydroxylase (C3′H), caffeoyl shikimate esterase (CSE), coumarate 3-hydroxylase (C3H), caffeic acid/5-hydroxyconiferaldehyde 3/5-O-methyltransferase (COMT), 4-hydroxycinnamate CoA ligase (4CL), caffeoyl CoA 3-O-methyltransferase (CCoAOMT), cinnamoyl CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), ferulic acid/coniferaldehyde 5-hydroxylase (F5H), laccase (LAC)
sign. The 3-OH group in caffeyl alcohol can be introduced by the direct action of C3H on coumarate, or by the combined activities of HCT, C3′H and CSE in the so-called “shikimate shunt”. S lignin is not made in the Cleome seed coat due to lack of expression of F5H. Enzymes are L-phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT), coumaroyl shikimate 3′-hydroxylase (C3′H), caffeoyl shikimate esterase (CSE), coumarate 3-hydroxylase (C3H), caffeic acid/5-hydroxyconiferaldehyde 3/5-O-methyltransferase (COMT), 4-hydroxycinnamate CoA ligase (4CL), caffeoyl CoA 3-O-methyltransferase (CCoAOMT), cinnamoyl CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), ferulic acid/coniferaldehyde 5-hydroxylase (F5H), laccase (LAC)


 sign. B–E Monolignol compositions of transgenic lines determined by thioacidolysis. Data show % of H-lignin monomer (B), C-lignin monomer (C), G-lignin monomer (D) and S-lignin monomer (E). cGUS, GUS control in comt mutant background; RGUS, GUS control in R108 wild-type background. Stem, mature R108 stem. −/−, no transgenic event; −/+, transgenic roots only harboring ChLAC8 overexpression construct; ±, transgenic roots only harboring MtCOMT-MtCCoAOMT RNAi construct; +/+, transgenic roots harboring both MtCOMT-MtCCoAOMT RNAi and ChLAC8 overexpression constructs. CCL, RNAi construct for MtCOMT (C) and MtCCoAOMT (C) and overexpression construct for ChLAC8 (L). Data are means ± SD derived from three biological replicates
sign. B–E Monolignol compositions of transgenic lines determined by thioacidolysis. Data show % of H-lignin monomer (B), C-lignin monomer (C), G-lignin monomer (D) and S-lignin monomer (E). cGUS, GUS control in comt mutant background; RGUS, GUS control in R108 wild-type background. Stem, mature R108 stem. −/−, no transgenic event; −/+, transgenic roots only harboring ChLAC8 overexpression construct; ±, transgenic roots only harboring MtCOMT-MtCCoAOMT RNAi construct; +/+, transgenic roots harboring both MtCOMT-MtCCoAOMT RNAi and ChLAC8 overexpression constructs. CCL, RNAi construct for MtCOMT (C) and MtCCoAOMT (C) and overexpression construct for ChLAC8 (L). Data are means ± SD derived from three biological replicates

Similar articles
-
Enzymatic basis for C-lignin monomer biosynthesis in the seed coat of Cleome hassleriana.Plant J. 2019 Aug;99(3):506-520. doi: 10.1111/tpj.14340. Epub 2019 May 17. Plant J. 2019. PMID: 31002459
-
Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula.Proc Natl Acad Sci U S A. 2010 Oct 12;107(41):17803-8. doi: 10.1073/pnas.1012900107. Epub 2010 Sep 27. Proc Natl Acad Sci U S A. 2010. PMID: 20876124 Free PMC article.
-
Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity.Phytochemistry. 2002 Oct;61(3):221-94. doi: 10.1016/s0031-9422(02)00211-x. Phytochemistry. 2002. PMID: 12359514 Review.
-
Substrate Specificity of LACCASE8 Facilitates Polymerization of Caffeyl Alcohol for C-Lignin Biosynthesis in the Seed Coat of Cleome hassleriana.Plant Cell. 2020 Dec;32(12):3825-3845. doi: 10.1105/tpc.20.00598. Epub 2020 Oct 9. Plant Cell. 2020. PMID: 33037146 Free PMC article.
-
[Advances in research on lignin biosynthesis and its genetic engineering].Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2004 Aug;30(4):361-70. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2004. PMID: 15627683 Review. Chinese.
Cited by
-
Benzenoid Aromatics from Renewable Resources.Chem Rev. 2024 Oct 9;124(19):10701-10876. doi: 10.1021/acs.chemrev.4c00087. Epub 2024 Sep 17. Chem Rev. 2024. PMID: 39288258 Free PMC article. Review.
References
-
- Barsberg ST, Lee Y-I, Rasmussen HN. Development of C-lignin with G/S-lignin and lipids in orchid seed coats—an unexpected diversity exposed by ATR-FT-IR spectroscopy. Seed Sci Res. 2017;28:41–51. doi: 10.1017/S0960258517000344. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
