Diversifying maize genomic selection models
- PMID: 37309328
- PMCID: PMC10236107
- DOI: 10.1007/s11032-021-01221-4
Diversifying maize genomic selection models
Abstract
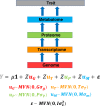
Genomic selection (GS) is one of the most powerful tools available for maize breeding. Its use of genome-wide marker data to estimate breeding values translates to increased genetic gains with fewer breeding cycles. In this review, we cover the history of GS and highlight particular milestones during its adaptation to maize breeding. We discuss how GS can be applied to developing superior maize inbreds and hybrids. Additionally, we characterize refinements in GS models that could enable the encapsulation of non-additive genetic effects, genotype by environment interactions, and multiple levels of the biological hierarchy, all of which could ultimately result in more accurate predictions of breeding values. Finally, we suggest the stages in a maize breeding program where it would be beneficial to apply GS. Given the current sophistication of high-throughput phenotypic, genotypic, and other -omic level data currently available to the maize community, now is the time to explore the implications of their incorporation into GS models and thus ensure that genetic gains are being achieved as quickly and efficiently as possible.
Keywords: GBLUP; Genomic selection; Hybrid prediction; Maize; Multi-kernel; Omics.
© The Author(s), under exclusive licence to Springer Nature B.V. 2021.
Conflict of interest statement
Conflict of interestThe authors declare no competing interests.
Figures
References
-
- Alliance G (2010) Genetics 101. Understanding genetics: a district of Colombia guide for patients and health professionals:22–32 https://www.resourcerepository.org/documents/1869/understandinggenetics:..., https://www.ncbi.nlm.nih.gov/books/NBK132149/pdf/Bookshelf_NBK132149.pdf
-
- Argueso CT, Assmann SM, Birnbaum KD, Chen S, Dinneny JR, Doherty CJ, Eveland AL, Friesner J, Greenlee VR, Law JA, Marshall-Colón A, Mason GA, O’Lexy R, Peck SC, Schmitz RJ, Song L, Stern D, Varagona MJ, Walley JW, Williams CM. Directions for research and training in plant omics: big questions and big data. Plant Direct. 2019;3(4):e00133. doi: 10.1002/pld3.133. - DOI - PMC - PubMed
-
- Arruda MP, Lipka AE, Brown PJ, Krill AM, Thurber C, Brown-Guedira G, Dong Y, Foresman BJ, Kolb FL. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.) Molecular Breeding. 2016;36(7):1–11. doi: 10.1007/s11032-016-0508-5. - DOI
LinkOut - more resources
Full Text Sources

