β-Catenin Drives Butyrophilin-like Molecule Loss and γδ T-cell Exclusion in Colon Cancer
- PMID: 37309673
- PMCID: PMC10398359
- DOI: 10.1158/2326-6066.CIR-22-0644
β-Catenin Drives Butyrophilin-like Molecule Loss and γδ T-cell Exclusion in Colon Cancer
Abstract
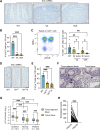
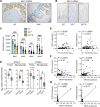
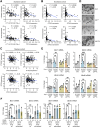
Intraepithelial lymphocytes (IEL) expressing γδ T-cell receptors (γδTCR) play key roles in elimination of colon cancer. However, the precise mechanisms by which progressing cancer cells evade immunosurveillance by these innate T cells are unknown. Here, we investigated how loss of the Apc tumor suppressor in gut tissue could enable nascent cancer cells to escape immunosurveillance by cytotoxic γδIELs. In contrast with healthy intestinal or colonic tissue, we found that γδIELs were largely absent from the microenvironment of both mouse and human tumors, and that butyrophilin-like (BTNL) molecules, which can critically regulate γδIEL through direct γδTCR interactions, were also downregulated in tumors. We then demonstrated that β-catenin activation through loss of Apc rapidly suppressed expression of the mRNA encoding the HNF4A and HNF4G transcription factors, preventing their binding to promoter regions of Btnl genes. Reexpression of BTNL1 and BTNL6 in cancer cells increased γδIEL survival and activation in coculture assays but failed to augment their cancer-killing ability in vitro or their recruitment to orthotopic tumors. However, inhibition of β-catenin signaling via genetic deletion of Bcl9/Bcl9L in either Apc-deficient or mutant β-catenin mouse models restored Hnf4a, Hnf4g, and Btnl gene expression and γδ T-cell infiltration into tumors. These observations highlight an immune-evasion mechanism specific to WNT-driven colon cancer cells that disrupts γδIEL immunosurveillance and furthers cancer progression.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- A25142/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_21042/MRC_/Medical Research Council/United Kingdom
- 29799/CRUK_/Cancer Research UK/United Kingdom
- FC001093/WT_/Wellcome Trust/United Kingdom
- 206314/WT_/Wellcome Trust/United Kingdom
- A29799/CRUK_/Cancer Research UK/United Kingdom
- 208990/WT_/Wellcome Trust/United Kingdom
- R01 DK121915/DK/NIDDK NIH HHS/United States
- 206314/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- FC001093/CRUK_/Cancer Research UK/United Kingdom
- R01 CA190558/CA/NCI NIH HHS/United States
- A21139/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

