A unique mRNA decapping complex in trypanosomes
- PMID: 37309887
- PMCID: PMC10415143
- DOI: 10.1093/nar/gkad497
A unique mRNA decapping complex in trypanosomes
Abstract
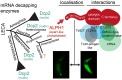
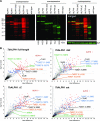
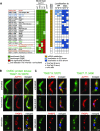
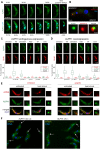
Removal of the mRNA 5' cap primes transcripts for degradation and is central for regulating gene expression in eukaryotes. The canonical decapping enzyme Dcp2 is stringently controlled by assembly into a dynamic multi-protein complex together with the 5'-3'exoribonuclease Xrn1. Kinetoplastida lack Dcp2 orthologues but instead rely on the ApaH-like phosphatase ALPH1 for decapping. ALPH1 is composed of a catalytic domain flanked by C- and N-terminal extensions. We show that T. brucei ALPH1 is dimeric in vitro and functions within a complex composed of the trypanosome Xrn1 ortholog XRNA and four proteins unique to Kinetoplastida, including two RNA-binding proteins and a CMGC-family protein kinase. All ALPH1-associated proteins share a unique and dynamic localization to a structure at the posterior pole of the cell, anterior to the microtubule plus ends. XRNA affinity capture in T. cruzi recapitulates this interaction network. The ALPH1 N-terminus is not required for viability in culture, but essential for posterior pole localization. The C-terminus, in contrast, is required for localization to all RNA granule types, as well as for dimerization and interactions with XRNA and the CMGC kinase, suggesting possible regulatory mechanisms. Most significantly, the trypanosome decapping complex has a unique composition, differentiating the process from opisthokonts.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

