The Molecular Phenotype of Kidney Transplants: Insights From the MMDx Project
- PMID: 37310258
- PMCID: PMC10718223
- DOI: 10.1097/TP.0000000000004624
The Molecular Phenotype of Kidney Transplants: Insights From the MMDx Project
Abstract
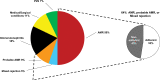
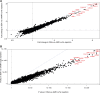
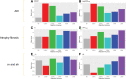
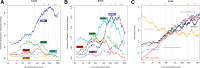
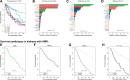
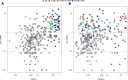
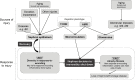
This review outlines the molecular disease states in kidney transplant biopsies as documented in the development of the Molecular Microscope Diagnostic System (MMDx). These states include T cell-mediated rejection (TCMR), antibody-mediated rejection (AMR), recent parenchymal injury, and irreversible atrophy-fibrosis. The MMDx project, initiated through a Genome Canada grant, is a collaboration involving many centers. MMDx uses genome-wide microarrays to measure transcript expression, interprets the results using ensembles of machine learning algorithms, and generates a report. Experimental studies in mouse models and cell lines were extensively used to annotate molecular features and interpret the biopsy results. Over time, MMDx revealed unexpected aspects of the disease states: for example, AMR is usually C4d-negative and often DSA-negative, and subtle "Minor" AMR-like states are frequent. Parenchymal injury correlates with both reduced glomerular filtration rate and increased risk of graft loss. In kidneys with rejection, injury features, not rejection activity, are the strongest predictors of graft survival. Both TCMR and AMR produce injury, but TCMR induces immediate nephron injury and accelerates atrophy-fibrosis, whereas AMR induces microcirculation and glomerular damage that slowly leads to nephron failure and atrophy-fibrosis. Plasma donor-derived cell-free DNA levels correlate strongly with AMR activity, acute kidney injury, and in a complex way with TCMR activity. Thus, the MMDx project has documented the molecular processes that underlie the clinical and histologic states in kidney transplants, and provides a diagnostic tool that can be used to calibrate biomarkers, optimize histology interpretation, and guide clinical trials.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc.
Figures



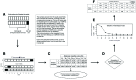
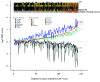


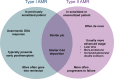

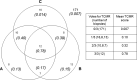

References
-
- Hariharan S, Israni AK, Danovitch G. Long-term survival after kidney transplantation. N Engl J Med. 2021;385:729–743. - PubMed
-
- Nankivell BJ, Kuypers DR. Diagnosis and prevention of chronic kidney allograft loss. Lancet. 2011;378:1428–1437. - PubMed
-
- Tait BD, Susal C, Gebel HM, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. 2013;95:19–47. - PubMed
-
- Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. - PubMed
-
- Xiao H, Gao F, Pang Q, et al. Diagnostic Accuracy of donor-derived cell-free DNA in renal-allograft rejection: a meta-analysis. Transplantation. 2021;105:1303–1310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

