Identification of novel genes associated with exercise and calorie restriction effects in skeletal muscle
- PMID: 37310402
- PMCID: PMC10292903
- DOI: 10.18632/aging.204793
Identification of novel genes associated with exercise and calorie restriction effects in skeletal muscle
Abstract
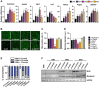
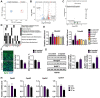
Exercise and caloric restriction (CR) significantly increase longevity across a range of species and delay aging-related losses in organ function. Although both interventions enhance skeletal muscle function, the molecular mechanisms underlying these associations are unknown. We sought to identify genes regulated by CR and exercise in muscle, and investigate their relationship with muscle function. To do this, expression profiles of Gene Expression Omnibus datasets obtained from the muscle tissue of calorie-restricted male primates and young men post-exercise were analyzed. There were seven transcripts (ADAMTS1, CPEB4, EGR2, IRS2, NR4A1, PYGO1, and ZBTB43) that were consistently upregulated by both CR and exercise training. We used C2C12 murine myoblasts to investigate the effect of silencing these genes on myogenesis, mitochondrial respiration, autophagy, and insulin signaling, all of which are processes affected by CR and exercise. Our results show that in C2C12 cells, Irs2 and Nr4a1 expression were critical for myogenesis, and five genes (Egr2, Irs2, Nr4a1, Pygo1, and ZBTB43) regulated mitochondrial respiration while having no effect on autophagy. Cpeb4 knockdown increased the expression of genes involved in muscle atrophy and induced myotube atrophy. These findings suggest new resources for studying the mechanisms underlying the beneficial effects of exercise and calorie restriction on skeletal muscle function and lifespan extension.
Keywords: calorie restriction; exercise; insulin sensitivity; mitochondrial respiration; muscle atrophy; myogenesis.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

