Isolation and Characterization of Porcine Endocardial Endothelial Cells
- PMID: 37310900
- PMCID: PMC10442675
- DOI: 10.1089/ten.TEC.2023.0009
Isolation and Characterization of Porcine Endocardial Endothelial Cells
Abstract
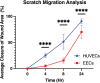
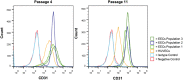
The heart contains diverse endothelial cell types. We sought to characterize the endocardial endothelial cells (EECs), which line the chambers of the heart. EECs are relatively understudied, yet their dysregulation can lead to various cardiac pathologies. Due to the lack of commercial availability of these cells, we reported our protocol for isolating EECs from porcine hearts and for establishing an EEC population through cell sorting. In addition, we compared the EEC phenotype and fundamental behaviors to a well-studied endothelial cell line, human umbilical vein endothelial cells (HUVECs). The EECs stained positively for classic phenotypic markers such as CD31, von Willebrand Factor, and vascular endothelial (VE) cadherin. The EECs proliferated more quickly than HUVECs at 48 h (1310 ± 251 cells vs. 597 ± 130 cells, p = 0.0361) and at 96 h (2873 ± 257 cells vs. 1714 ± 342 cells, p = 0.0002). Yet EECs migrated more slowly than HUVECs to cover a scratch wound at 4 h (5% ± 1% wound closure vs. 25% ± 3% wound closure, p < 0.0001), 8 h (15% ± 4% wound closure vs. 51% ± 12% wound closure, p < 0.0001), and 24 h (70% ± 11% wound closure vs. 90% ± 3% wound closure, p < 0.0001). Finally, the EECs maintained their endothelial phenotype by positive expression of CD31 through more than a dozen passages (three populations of EECs showing 97% ± 1% CD31+ cells in over 14 passages). In contrast, the HUVECs showed significantly reduced CD31 expression over high passages (80% ± 11% CD31+ cells over 14 passages). These important phenotypic differences between EECs and HUVECs highlight the need for researchers to utilize the most relevant cell types when studying or modeling diseases of interest.
Keywords: behavior; cell isolation; endocardial; endothelial cells; phenotype.
Conflict of interest statement
No competing financial interests exist.
Figures





Similar articles
-
Antithrombotic effects of endocardial endothelial cells-comparison with coronary artery endothelial cells.Prostaglandins. 1997 May;53(5):305-19. doi: 10.1016/0090-6980(97)00039-7. Prostaglandins. 1997. PMID: 9247971
-
Isolation of Endocardial and Coronary Endothelial Cells from the Ventricular Free Wall of the Rat Heart.J Vis Exp. 2020 Apr 15;(158). doi: 10.3791/61126. J Vis Exp. 2020. PMID: 32364545
-
The effects of transmural pressure on prostacyclin release from porcine endocardial endothelial cells--comparison with vascular endothelial cells.Pflugers Arch. 1997 Apr;433(6):848-50. doi: 10.1007/PL00008078. Pflugers Arch. 1997. PMID: 9049180
-
Roles of nuclear NPY and NPY receptors in the regulation of the endocardial endothelium and heart function.Can J Physiol Pharmacol. 2006 Jul;84(7):695-705. doi: 10.1139/y05-162. Can J Physiol Pharmacol. 2006. PMID: 16998533 Review.
-
Neuropeptide Y and its receptors in ventricular endocardial endothelial cells.Can J Physiol Pharmacol. 2017 Oct;95(10):1224-1229. doi: 10.1139/cjpp-2017-0290. Epub 2017 Jul 24. Can J Physiol Pharmacol. 2017. PMID: 28738162 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

