Reference-based cell type matching of in situ image-based spatial transcriptomics data on primary visual cortex of mouse brain
- PMID: 37311768
- PMCID: PMC10264402
- DOI: 10.1038/s41598-023-36638-8
Reference-based cell type matching of in situ image-based spatial transcriptomics data on primary visual cortex of mouse brain
Abstract
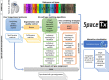
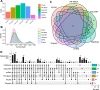
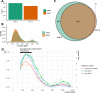
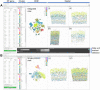
With the advent of multiplex fluorescence in situ hybridization (FISH) and in situ RNA sequencing technologies, spatial transcriptomics analysis is advancing rapidly, providing spatial location and gene expression information about cells in tissue sections at single cell resolution. Cell type classification of these spatially-resolved cells can be inferred by matching the spatial transcriptomics data to reference atlases derived from single cell RNA-sequencing (scRNA-seq) in which cell types are defined by differences in their gene expression profiles. However, robust cell type matching of the spatially-resolved cells to reference scRNA-seq atlases is challenging due to the intrinsic differences in resolution between the spatial and scRNA-seq data. In this study, we systematically evaluated six computational algorithms for cell type matching across four image-based spatial transcriptomics experimental protocols (MERFISH, smFISH, BaristaSeq, and ExSeq) conducted on the same mouse primary visual cortex (VISp) brain region. We find that many cells are assigned as the same type by multiple cell type matching algorithms and are present in spatial patterns previously reported from scRNA-seq studies in VISp. Furthermore, by combining the results of individual matching strategies into consensus cell type assignments, we see even greater alignment with biological expectations. We present two ensemble meta-analysis strategies used in this study and share the consensus cell type matching results in the Cytosplore Viewer ( https://viewer.cytosplore.org ) for interactive visualization and data exploration. The consensus matching can also guide spatial data analysis using SSAM, allowing segmentation-free cell type assignment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Computational solutions for spatial transcriptomics.Comput Struct Biotechnol J. 2022 Sep 1;20:4870-4884. doi: 10.1016/j.csbj.2022.08.043. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147664 Free PMC article. Review.
-
Concordance of MERFISH spatial transcriptomics with bulk and single-cell RNA sequencing.Life Sci Alliance. 2022 Dec 16;6(1):e202201701. doi: 10.26508/lsa.202201701. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36526371 Free PMC article.
-
Cell segmentation-free inference of cell types from in situ transcriptomics data.Nat Commun. 2021 Jun 10;12(1):3545. doi: 10.1038/s41467-021-23807-4. Nat Commun. 2021. PMID: 34112806 Free PMC article.
-
MLSpatial: A machine-learning method to reconstruct the spatial distribution of cells from scRNA-seq by extracting spatial features.Comput Biol Med. 2023 Jun;159:106873. doi: 10.1016/j.compbiomed.2023.106873. Epub 2023 Apr 18. Comput Biol Med. 2023. PMID: 37105115
-
Deep learning in integrating spatial transcriptomics with other modalities.Brief Bioinform. 2024 Nov 22;26(1):bbae719. doi: 10.1093/bib/bbae719. Brief Bioinform. 2024. PMID: 39800876 Free PMC article. Review.
Cited by
-
Challenges and best practices in omics benchmarking.Nat Rev Genet. 2024 May;25(5):326-339. doi: 10.1038/s41576-023-00679-6. Epub 2024 Jan 12. Nat Rev Genet. 2024. PMID: 38216661 Review.
-
Modular strategies for spatial mapping of diverse cell type data of the mouse brain.Res Sq [Preprint]. 2025 Apr 9:rs.3.rs-6289741. doi: 10.21203/rs.3.rs-6289741/v1. Res Sq. 2025. PMID: 40297692 Free PMC article. Preprint.
-
High-resolution mapping of single cells in spatial context.Nat Commun. 2025 Jul 15;16(1):6533. doi: 10.1038/s41467-025-61667-4. Nat Commun. 2025. PMID: 40664667 Free PMC article.
-
SpaceWalker enables interactive gradient exploration for spatial transcriptomics data.Cell Rep Methods. 2023 Dec 18;3(12):100645. doi: 10.1016/j.crmeth.2023.100645. Epub 2023 Nov 15. Cell Rep Methods. 2023. PMID: 37972590 Free PMC article.
-
Optimizing Xenium In Situ data utility by quality assessment and best-practice analysis workflows.Nat Methods. 2025 Apr;22(4):813-823. doi: 10.1038/s41592-025-02617-2. Epub 2025 Mar 13. Nat Methods. 2025. PMID: 40082609 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

