Quantum chemical data generation as fill-in for reliability enhancement of machine-learning reaction and retrosynthesis planning
- PMID: 37312681
- PMCID: PMC10259370
- DOI: 10.1039/d3dd00006k
Quantum chemical data generation as fill-in for reliability enhancement of machine-learning reaction and retrosynthesis planning
Abstract
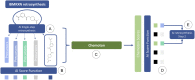
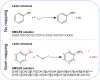
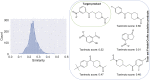
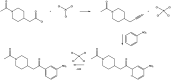
Data-driven synthesis planning has seen remarkable successes in recent years by virtue of modern approaches of artificial intelligence that efficiently exploit vast databases with experimental data on chemical reactions. However, this success story is intimately connected to the availability of existing experimental data. It may well occur in retrosynthetic and synthesis design tasks that predictions in individual steps of a reaction cascade are affected by large uncertainties. In such cases, it will, in general, not be easily possible to provide missing data from autonomously conducted experiments on demand. However, first-principles calculations can, in principle, provide missing data to enhance the confidence of an individual prediction or for model retraining. Here, we demonstrate the feasibility of such an ansatz and examine resource requirements for conducting autonomous first-principles calculations on demand.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


References
LinkOut - more resources
Full Text Sources
