The complete mitochondrial genome of Stibochiona nicea (Gray, 1846) (Lepidoptera: Nymphalidae) and phylogenetic analysis
- PMID: 37312971
- PMCID: PMC10259338
- DOI: 10.1080/23802359.2023.2221348
The complete mitochondrial genome of Stibochiona nicea (Gray, 1846) (Lepidoptera: Nymphalidae) and phylogenetic analysis
Abstract
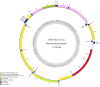
In this study, the complete mitochondrial genome (mitogenome) of Stibochiona nicea (Gray, 1846) (Lepidoptera: Nymphalidae) was first reported with 15,298 bp in size, containing 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes (rrnL and rrnS), and one control region. The nucleotide composition of the entire mitogenome is highly A + T biased (81.5%). The gene content and arrangement of the newly sequenced mitogenome are identical to those of the other available mitogenomes of Nymphalidae. All PCGs start with the conventional ATN codons, except for cox1 initiating with atypical CGA(R). Nine PCGs (atp8, atp6, cox3, nad1, nad2, nad3, nad4l, nad6, and cob) utilize a typical stop codon TAA, whereas the remaining PCGs (cox1, cox2, nad4, and nad5) end with an incomplete stop codon T-. Phylogenetic analysis revealed that S. nicea is closely related to Dichorragia nesimachus within Pseudergolinae, which further forms the sister group to the grouping of (Nymphalinae + (Cyrestinae + (Biblidinae + Apaturinae))). The complete mitogenome of S. nicea will provide useful genetic information for improving the taxonomic system and phylogenetics of Nymphalidae.
Keywords: Nymphalidae; Stibochiona nicea; mitochondrial genome; phylogeny.
© 2023 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Ackery PR, Jong R, Vane-Wright RI.. 1999. The butterflies: Hedyloidea, Hesperioidea, and Papilionoidea. In: Kristensen NP, editor. Lepidoptera: moths and butterflies. 1. Evolution, systematics, and biogeography. Handbook of zoology. Vol. IV,Part35. New York: de Gruyter; p. 263–299.
-
- Boggs CL, Watt WB, Ehrlich PR.. 2003. Butterflies: evolution and ecology taking flight. Chicago, IL: University of Chicago Press.
-
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117. - PubMed
-
- Espeland M, Breinholt J, Willmott KR, Warren AD, Vila R, Toussaint EFA, Maunsell SC, Aduse-Poku K, Talavera G, Eastwood R, et al. 2018. A comprehensive and dated phylogenomic analysis of butterflies. Curr Biol. 28(5):770–778.e5. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
