Characterization of the complete chloroplast genome of Gypsophila huashanensis Y. W. Tsui & D. Q. Lu, an endemic herb species in China
- PMID: 37312972
- PMCID: PMC10259312
- DOI: 10.1080/23802359.2023.2220436
Characterization of the complete chloroplast genome of Gypsophila huashanensis Y. W. Tsui & D. Q. Lu, an endemic herb species in China
Abstract
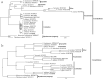
Gypsophila huashanensis Y. W. Tsui & D. Q. Lu (Caryophyllaceae) is an endemic herb species to the Qinling Mountains in China. In this study, we characterized its whole plastid genome using the Illumina sequencing platform. The complete plastid genome of G. huashanensis is 152,457 bp in length, including a large single-copy DNA region of 83,476 bp, a small single-copy DNA region of 17,345 bp, and a pair of inverted repeat DNA sequences of 25,818 bp. The genome contains 130 genes comprising 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Evolutionary analysis showed that the non-coding regions of Caryophyllaceae exhibit a higher level of divergence than the exon regions. Gene site selection analysis suggested that 11 coding protein genes (accD, atpF, ndhA, ndhB, petB, petD, rpoCl, rpoC2, rps16, ycfl, and ycf2) have some sites under protein sequence evolution. Phylogenetic analysis showed that G. huashanensis is most closely related to the congeneric species G. oldhamiana. These results are very useful for studying phylogenetic evolution and species divergence in the family Caryophyllaceae.
Keywords: Chloroplast genome; Gypsophila huashanensis; evolutionary selection; phylogenetic relationship.
© 2023 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Doyle JJ, Doyle JL.. 1990. Isolation of plant DNA from plant tissue. Focus. 12:13–15.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
