Machine learning analysis of humoral and cellular responses to SARS-CoV-2 infection in young adults
- PMID: 37313411
- PMCID: PMC10258347
- DOI: 10.3389/fimmu.2023.1158905
Machine learning analysis of humoral and cellular responses to SARS-CoV-2 infection in young adults
Abstract
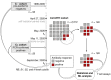
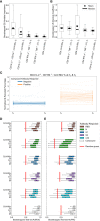
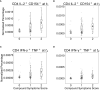
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces B and T cell responses, contributing to virus neutralization. In a cohort of 2,911 young adults, we identified 65 individuals who had an asymptomatic or mildly symptomatic SARS-CoV-2 infection and characterized their humoral and T cell responses to the Spike (S), Nucleocapsid (N) and Membrane (M) proteins. We found that previous infection induced CD4 T cells that vigorously responded to pools of peptides derived from the S and N proteins. By using statistical and machine learning models, we observed that the T cell response highly correlated with a compound titer of antibodies against the Receptor Binding Domain (RBD), S and N. However, while serum antibodies decayed over time, the cellular phenotype of these individuals remained stable over four months. Our computational analysis demonstrates that in young adults, asymptomatic and paucisymptomatic SARS-CoV-2 infections can induce robust and long-lasting CD4 T cell responses that exhibit slower decays than antibody titers. These observations imply that next-generation COVID-19 vaccines should be designed to induce stronger cellular responses to sustain the generation of potent neutralizing antibodies.
Keywords: SARS-CoV-2; T cell response; antibody titers; machine learning; neutralizing antibodies.
Copyright © 2023 Marcinkevics, Silva, Hankele, Dörnte, Kadelka, Csik, Godbersen, Goga, Hasenöhrl, Hirschi, Kabakci, LaPierre, Mayrhofer, Title, Shu, Baiioud, Bernal, Dassisti, Saenz-de-Juano, Schmidhauser, Silvestrelli, Ulbrich, Ulbrich, Wyss, Stekhoven, Al-Quaddoomi, Yu, Binder, Schultheiβ, Zindel, Kolling, Goldhahn, Seighalani, Zjablovskaja, Hardung, Schuster, Richter, Huang, Lauer, Baurmann, Low, Vaqueirinho, Jovic, Piccoli, Ciesek, Vogt, Sallusto, Stoffel and Ulbrich.
Conflict of interest statement
CD, MSc, BS, PZ, FH, AR, Y-JH, GL, and HB are employees of Miltenyi Biotec B.V. & Co. KG. LP is an employee of Humabs BioMed SA, a subsidiary of Vir Biotechnology. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

