Towards understanding paleoclimate impacts on primate de novo genes
- PMID: 37313728
- PMCID: PMC10468307
- DOI: 10.1093/g3journal/jkad135
Towards understanding paleoclimate impacts on primate de novo genes
Abstract
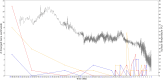
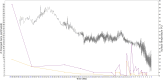
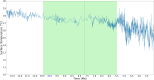
De novo genes are genes that emerge as new genes in some species, such as primate de novo genes that emerge in certain primate species. Over the past decade, a great deal of research has been conducted regarding their emergence, origins, functions, and various attributes in different species, some of which have involved estimating the ages of de novo genes. However, limited by the number of species available for whole-genome sequencing, relatively few studies have focused specifically on the emergence time of primate de novo genes. Among those, even fewer investigate the association between primate gene emergence with environmental factors, such as paleoclimate (ancient climate) conditions. This study investigates the relationship between paleoclimate and human gene emergence at primate species divergence. Based on 32 available primate genome sequences, this study has revealed possible associations between temperature changes and the emergence of de novo primate genes. Overall, findings in this study are that de novo genes tended to emerge in the recent 13 MY when the temperature continues cooling, which is consistent with past findings. Furthermore, in the context of an overall trend of cooling temperature, new primate genes were more likely to emerge during local warming periods, where the warm temperature more closely resembled the environmental condition that preceded the cooling trend. Results also indicate that both primate de novo genes and human cancer-associated genes have later origins in comparison to random human genes. Future studies can be in-depth on understanding human de novo gene emergence from an environmental perspective as well as understanding species divergence from a gene emergence perspective.
Keywords: de novo genes; gene emergence; gene evolution; paleoclimate; primate genes.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures





References
-
- Badgley C, Gingerich PD. Sampling and faunal turnover in early Eocene mammals. Palaeogeogr Palaeoclimatol Palaeoecol. 1988;63:141–157. doi:10.1016/0031-0182(88)90094-6 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous
