Massive and Lengthy Clonal Nosocomial Expansion of Mycobacterium abscessus subsp. massiliense among Patients Who Are Ventilator Dependent without Cystic Fibrosis
- PMID: 37314340
- PMCID: PMC10433864
- DOI: 10.1128/spectrum.04908-22
Massive and Lengthy Clonal Nosocomial Expansion of Mycobacterium abscessus subsp. massiliense among Patients Who Are Ventilator Dependent without Cystic Fibrosis
Abstract
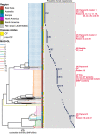
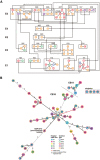
Nontuberculous mycobacterial infections are generally believed to be independently acquired from the environment. Although person-to-person transmission of nontuberculous mycobacteria, especially Mycobacterium abscessus subsp. massiliense, is a serious concern among individuals with cystic fibrosis (CF), evidence of its spread among patients without CF has never been established. We unexpectedly found a number of M. abscessus subsp. massiliense cases among patients without CF in a hospital. This study aimed to define the mechanism of M. abscessus subsp. massiliense infection among patients who were ventilator dependent and without CF who had progressive neurodegenerative diseases in our long-term care wards from 2014 to 2018 during suspected nosocomial outbreaks. We conducted whole-genome sequencing of M. abscessus subsp. massiliense isolates from 52 patients and environmental samples. Potential opportunities for in-hospital transmission were analyzed using epidemiological data. M. abscessus subsp. massiliense was isolated from one air sample obtained near a patient without CF who was colonized with M. abscessus subsp. massiliense but not from other potential sources. Phylogenetic analysis of the strains from these patients and the environmental isolate revealed clonal expansion of near-identical M. abscessus subsp. massiliense isolates, with the isolates generally differing by fewer than 22 single nucleotide polymorphisms (SNPs). Approximately half of the isolates differed by fewer than nine SNPs, indicating interpatient transmission. Whole-genome sequencing revealed a potential nosocomial outbreak among patients who were ventilator dependent and without CF. IMPORTANCE The isolation of M. abscessus subsp. massiliense from the air, but not from environmental fluid samples, may suggest airborne transmission. This was the first report to demonstrate person-to-person transmission of M. abscessus subsp. massiliense, even among patients without CF. M. abscessus subsp. massiliense may spread among patients who are ventilator dependent without CF through direct or indirect in-hospital transmission. The current infection control measures should address potential transmission among patients without CF, particularly in facilities that treat patients who are ventilator dependent and patients with preexisting chronic pulmonary diseases, such as CF.
Keywords: Mycobacterium abscessus subsp. massiliense; nontuberculous mycobacteria; progressive neurodegenerative disease; transmission; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Genomic Analysis of Mycobacterium abscessus Complex Isolates Collected in Ireland between 2006 and 2017.J Clin Microbiol. 2020 Jun 24;58(7):e00295-20. doi: 10.1128/JCM.00295-20. Print 2020 Jun 24. J Clin Microbiol. 2020. PMID: 32295892 Free PMC article.
-
Potential Cross-Transmission of Mycobacterium abscessus among Non-Cystic Fibrosis Patients at a Tertiary Hospital in Japan.Microbiol Spectr. 2022 Jun 29;10(3):e0009722. doi: 10.1128/spectrum.00097-22. Epub 2022 May 10. Microbiol Spectr. 2022. PMID: 35536059 Free PMC article.
-
Molecular Epidemiology of Mycobacterium abscessus Isolates Recovered from German Cystic Fibrosis Patients.Microbiol Spectr. 2022 Aug 31;10(4):e0171422. doi: 10.1128/spectrum.01714-22. Epub 2022 Aug 8. Microbiol Spectr. 2022. PMID: 35938728 Free PMC article.
-
Mycobacterium abscessus. "Pleased to meet you, hope you guess my name...".Ann Am Thorac Soc. 2015 Mar;12(3):436-9. doi: 10.1513/AnnalsATS.201501-015OI. Ann Am Thorac Soc. 2015. PMID: 25643064 Review.
-
Mycobacterium abscessus: insights from a bioinformatic perspective.Crit Rev Microbiol. 2023 Aug;49(4):499-514. doi: 10.1080/1040841X.2022.2082268. Epub 2022 Jun 13. Crit Rev Microbiol. 2023. PMID: 35696783 Review.
Cited by
-
Effects of Amikacin Liposome Inhalation Suspension and Amikacin Resistance Development in Patients With Refractory Mycobacterium avium Complex Pulmonary Disease.Open Forum Infect Dis. 2025 Mar 1;12(3):ofaf118. doi: 10.1093/ofid/ofaf118. eCollection 2025 Mar. Open Forum Infect Dis. 2025. PMID: 40110422 Free PMC article.
-
Identifying healthcare transmission routes of nontuberculous mycobacteria with whole genome sequencing: a systematic review.Infect Control Hosp Epidemiol. 2025 Feb 3;46(4):1-6. doi: 10.1017/ice.2025.6. Online ahead of print. Infect Control Hosp Epidemiol. 2025. PMID: 39895079 Free PMC article.
-
High rate of macrolide resistance and closely genetically related Mycobacterium abscessus complex strains identified among both cystic fibrosis and non-cystic fibrosis patients within two countries.Microbiol Spectr. 2024 Oct 23;12(12):e0105624. doi: 10.1128/spectrum.01056-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39440987 Free PMC article.
-
Infection by Clonally Related Mycobacterium abscessus Isolates: The Role of Drinking Water.Am J Respir Crit Care Med. 2025 May;211(5):842-853. doi: 10.1164/rccm.202409-1824OC. Am J Respir Crit Care Med. 2025. PMID: 40072241
-
Whole-genome recombination and dynamic accessory genomes drive the phenotypic diversity of Mycobacterium abscessus subspecies.Ann Clin Microbiol Antimicrob. 2025 Jul 22;24(1):44. doi: 10.1186/s12941-025-00804-9. Ann Clin Microbiol Antimicrob. 2025. PMID: 40696396 Free PMC article.
References
-
- Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann JL, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, Hempstead SE, Koetz K, Sabadosa KA, Sermet-Gaudelus I, Smyth AR, van Ingen J, Wallace RJ, Winthrop KL, Marshall BC, Haworth CS, US Cystic Fibrosis Foundation, European Cystic Fibrosis Society . 2016. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis. Thorax 71(Suppl 1):i1–i22. doi:10.1136/thoraxjnl-2015-207360. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

