Optimized single-cell RNA sequencing protocol to study early genome activation in mammalian preimplantation development
- PMID: 37314922
- PMCID: PMC10277609
- DOI: 10.1016/j.xpro.2023.102357
Optimized single-cell RNA sequencing protocol to study early genome activation in mammalian preimplantation development
Abstract
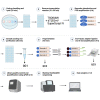
Here, we present a modification of single-cell tagged reverse transcription protocol to study gene expression on a single-cell level or with limited RNA input. We describe different enzymes for reverse transcription and cDNA amplification, modified lysis buffer, and additional clean-up steps before cDNA amplification. We also detail an optimized single-cell RNA sequencing method for handpicked single cells, or tens to hundreds of cells, as input material to study mammalian preimplantation development. For complete details on the use and execution of this protocol, please refer to Ezer et al.1.
Keywords: Bioinformatics; Developmental biology; Gene Expression; Genomics; Molecular Biology; RNA-seq; Sequence analysis; Sequencing; Single Cell.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

