GRAd-COV2 vaccine provides potent and durable humoral and cellular immunity to SARS-CoV-2 in randomized placebo-controlled phase 2 trial
- PMID: 37315558
- PMCID: PMC10243192
- DOI: 10.1016/j.xcrm.2023.101084
GRAd-COV2 vaccine provides potent and durable humoral and cellular immunity to SARS-CoV-2 in randomized placebo-controlled phase 2 trial
Abstract
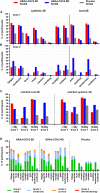
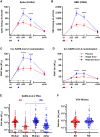
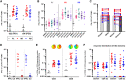
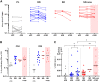
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic and heterologous immunization approaches implemented worldwide for booster doses call for diversified vaccine portfolios. GRAd-COV2 is a gorilla adenovirus-based COVID-19 vaccine candidate encoding prefusion-stabilized spike. The safety and immunogenicity of GRAd-COV2 is evaluated in a dose- and regimen-finding phase 2 trial (COVITAR study, ClinicalTrials.gov: NCT04791423) whereby 917 eligible participants are randomized to receive a single intramuscular GRAd-COV2 administration followed by placebo, or two vaccine injections, or two doses of placebo, spaced over 3 weeks. Here, we report that GRAd-COV2 is well tolerated and induces robust immune responses after a single immunization; a second administration increases binding and neutralizing antibody titers. Potent, variant of concern (VOC) cross-reactive spike-specific T cell response peaks after the first dose and is characterized by high frequencies of CD8s. T cells maintain immediate effector functions and high proliferative potential over time. Thus, GRAd vector is a valuable platform for genetic vaccine development, especially when robust CD8 response is needed.
Keywords: CD4; CD8; COVID-19; Sars-CoV-2 vaccine; T cell response; immunological memory; neutralizing antibodies; phase 2 clinical trial; safety; simian adenoviral vector.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S. Capone, R. Camerini, R.D., F.G., S. Battella, A.M.C., G.P., S. Colloca, and A.F. are full employees of ReiThera Srl. S. Colloca and A.F. are founders and shareholders of Keires AG. S. Colloca is named inventor of the patent application no. 20183515.4 titled “GORILLA ADENOVIRUS NUCLEIC ACID- AND AMINO ACID-SEQUENCES, VECTORS CONTAINING SAME, AND USES THEREOF.” L.V. is full employee of Exom, the CRO in charge of the COVITAR study management. S.L.C. received honoraria from Gilead, ViiV, GSK, Janssen, and MSD, has participated on the advisory boards of Gilead, ViiV, GSK, Janssen, and MSD, and has received support for attending meetings from Gilead. M. Lichtner received honoraria and support for attending meetings from Gilead, MSD, and ViiV, participated on the advisory boards of ViiV, Abbvie, and MSD, and received grants through the institution from Gilead and Abbvie. R. Carsetti was a member of the COVITAR study steering committee. C.I. received financial support from Exom for statistical analysis of the COVITAR study.
Figures

References
-
- Medicine L.S.o.H.T. https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/
-
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

