SINE RNA of the imprinted miRNA clusters mediates constitutive type III interferon expression and antiviral protection in hemochorial placentas
- PMID: 37315561
- PMCID: PMC10524649
- DOI: 10.1016/j.chom.2023.05.018
SINE RNA of the imprinted miRNA clusters mediates constitutive type III interferon expression and antiviral protection in hemochorial placentas
Erratum in
-
SINE RNA of the imprinted miRNA clusters mediates constitutive type III interferon expression and antiviral protection in hemochorial placentas.Cell Host Microbe. 2024 Nov 13;32(11):2035. doi: 10.1016/j.chom.2024.10.003. Epub 2024 Oct 12. Cell Host Microbe. 2024. PMID: 39396520 Free PMC article. No abstract available.
Abstract
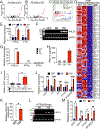
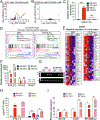
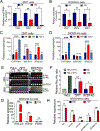
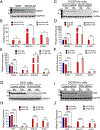
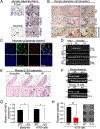
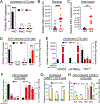
Hemochorial placentas have evolved defense mechanisms to prevent the vertical transmission of viruses to the immunologically underdeveloped fetus. Unlike somatic cells that require pathogen-associated molecular patterns to stimulate interferon production, placental trophoblasts constitutively produce type III interferons (IFNL) through an unknown mechanism. We demonstrate that transcripts of short interspersed nuclear elements (SINEs) embedded in miRNA clusters within the placenta trigger a viral mimicry response that induces IFNL and confers antiviral protection. Alu SINEs within primate-specific chromosome 19 (C19MC) and B1 SINEs within rodent-specific microRNA cluster on chromosome 2 (C2MC) produce dsRNAs that activate RIG-I-like receptors (RLRs) and downstream IFNL production. Homozygous C2MC knockout mouse trophoblast stem (mTS) cells and placentas lose intrinsic IFN expression and antiviral protection, whereas B1 RNA overexpression restores C2MCΔ/Δ mTS cell viral resistance. Our results uncover a convergently evolved mechanism whereby SINE RNAs drive antiviral resistance in hemochorial placentas, placing SINEs as integral players in innate immunity.
Keywords: C19MC; C2MC; antiviral immunity; convergent evolution; placenta; retrotransposons; vertical transmission.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.T.-J. is listed as inventor on patent application related to this project.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

