LC3B is lipidated to large lipid droplets during prolonged starvation for noncanonical autophagy
- PMID: 37315562
- PMCID: PMC10686041
- DOI: 10.1016/j.devcel.2023.05.009
LC3B is lipidated to large lipid droplets during prolonged starvation for noncanonical autophagy
Abstract
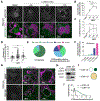
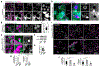
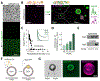
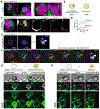
Lipid droplets (LDs) store lipids that can be utilized during times of scarcity via autophagic and lysosomal pathways, but how LDs and autophagosomes interact remained unclear. Here, we discovered that the E2 autophagic enzyme, ATG3, localizes to the surface of certain ultra-large LDs in differentiated murine 3T3-L1 adipocytes or Huh7 human liver cells undergoing prolonged starvation. Subsequently, ATG3 lipidates microtubule-associated protein 1 light-chain 3B (LC3B) to these LDs. In vitro, ATG3 could bind alone to purified and artificial LDs to mediate this lipidation reaction. We observed that LC3B-lipidated LDs were consistently in close proximity to collections of LC3B-membranes and were lacking Plin1. This phenotype is distinct from macrolipophagy, but it required autophagy because it disappeared following ATG5 or Beclin1 knockout. Our data suggest that extended starvation triggers a noncanonical autophagy mechanism, similar to LC3B-associated phagocytosis, in which the surface of large LDs serves as an LC3B lipidation platform for autophagic processes.
Keywords: Atg3; LC3B; lipid droplets; noncanonical autophagy; organelle biogenesis; prolonged starvation.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Mizushima N, and Komatsu M (2011). Autophagy: renovation of cells and tissues. Cell 147, 728–741. - PubMed
-
- Mizushima N, Ohsumi Y, and Yoshimori T (2002). Autophagosome formation in mammalian cells. Cell Struct. Funct 27, 421–429. - PubMed
-
- Mizushima N (2020). The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol 63, 1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

