Ubiquitin-like protein 5 is a novel player in the UPR-PERK arm and ER stress-induced cell death
- PMID: 37315790
- PMCID: PMC10339194
- DOI: 10.1016/j.jbc.2023.104915
Ubiquitin-like protein 5 is a novel player in the UPR-PERK arm and ER stress-induced cell death
Abstract
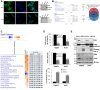
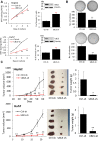
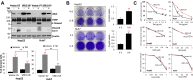
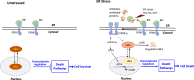
Biological functions of the highly conserved ubiquitin-like protein 5 (UBL5) are not well understood. In Caenorhabditis elegans, UBL5 is induced under mitochondrial stress to mount the mitochondrial unfolded protein response (UPR). However, the role of UBL5 in the more prevalent endoplasmic reticulum (ER) stress-UPR in the mammalian system is unknown. In the present work, we demonstrated that UBL5 was an ER stress-responsive protein, undergoing rapid depletion in mammalian cells and livers of mice. The ER stress-induced UBL5 depletion was mediated by proteasome-dependent yet ubiquitin-independent proteolysis. Activation of the protein kinase R-like ER kinase arm of the UPR was essential and sufficient for inducing UBL5 degradation. RNA-Seq analysis of UBL5-regulated transcriptome revealed that multiple death pathways were activated in UBL5-silenced cells. In agreement with this, UBL5 knockdown induced severe apoptosis in culture and suppressed tumorigenicity of cancer cells in vivo. Furthermore, overexpression of UBL5 protected specifically against ER stress-induced apoptosis. These results identify UBL5 as a physiologically relevant survival regulator that is proteolytically depleted by the UPR-protein kinase R-like ER kinase pathway, linking ER stress to cell death.
Keywords: ER stress; PERK; UBL5; UPR; apoptosis; cell survival; proteasome degradation; ubiquitin-independent proteasome system.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Clague M.J., Urbé S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. - PubMed
-
- Van Der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. - PubMed
-
- Collier G.R., McMillan J.S., Windmill K., Walder K., Tenne-Brown J., De Silva A., et al. Beacon: a novel gene involved in the regulation of energy balance. Diabetes. 2000;49:1766–1771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

